 3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual
3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual Results from an EMSL Arrows Calculation
| EMSL Arrows is a revolutionary approach to materials and chemical simulations that uses NWChem and chemical computational databases to make materials and chemical modeling accessible via a broad spectrum of digital communications including posts to web APIs, social networks, and traditional email. |
Molecular modeling software has previously been extremely complex, making it prohibative to all but experts in the field, yet even experts can struggle to perform calculations. This service is designed to be used by experts and non-experts alike. Experts can carry out and keep track of large numbers of complex calculations with diverse levels of theories present in their workflows. Additionally, due to a streamlined and easy-to-use input, non-experts can carry out a wide variety of molecular modeling calculations previously not accessible to them.
The id(s) for emsiles = Cl[C](Cl)Cl theory{pspw} xc{pbe} basis{100.0 Ry} solvation_type{None} ^{1} are: 63004
Use id=% instead of esmiles to print other entries.
mformula = C1Cl3
iupac = chloroform cation
PubChem = 6212
PubChem LCSS = 6212
cas = 67-66-3
kegg = C13827
synonyms = CHLOROFORM; Trichloromethane; 67-66-3; Methane, trichloro-; Trichlormethan; Formyl trichloride; Trichloroform; Methane trichloride; Methenyl trichloride; Methyl trichloride; Chloroforme; CHCl3; Cloroformio; Triclorometano; Trichloormethaan; Freon 20; R 20 (Refrigerant); 1,1,1-Trichloromethane; RCRA waste number U044; Methenyl chloride; NCI-C02686; Methylidyne trichloride; Refrigerant R20; HSDB 56; trichloro-methane; R 20; NSC 77361; UNII-7V31YC746X; Chloroform [UN1888] [Poison]; chloroform (trichloromethane); CHEBI:35255; 7V31YC746X; MFCD00000826; NCGC00090794-01; chloroform CHCl3; Chloroform, HPLC Grade; DSSTox_CID_306; DSSTox_RID_75501; DSSTox_GSID_20306; Chloroform, analytical standard; Chloroforme [French]; Caswell No. 192; Cloroformio [Italian]; Trichlormethan [Czech]; Chloroform [NF XVII]; Trichloormethaan [Dutch]; Triclorometano [Italian]; CAS-67-66-3; Chloroform [NF]; CCRIS 137; Chloroform, for HPLC, stabilized with amylene; CHLOROFORM, ACS; CHLOROFORMWith Amylene; CHLOROFORMWith Ethanol; EINECS 200-663-8; UN1888; Chloroform, 99+%, for HPLC, stabilized with ethanol; RCRA waste no. U044; Chloroform, 99+%, extra pure, stabilized with amylene; Chloroform, 99+%, extra pure, stabilized with ethanol; Chloroform, 99+%, for analysis, stabilized with ethanol; Chloroform, 99.9%, Extra Dry, stabilized, AcroSeal(R); EPA Pesticide Chemical Code 020701; BRN 1731042; chlorform; chloroforrn; cloroform; trichlormethane; trichlorocarbon; Chloroform, 99.8%, for analysis, stabilized with amylene; Chloroform, 99.8+%, ACS reagent, stabilized with ethanol; Chloroform, 99+%, for spectroscopy, stabilized with amylene; chloro form; chloro-form; Chloroform-; AI3-24207; Chloroform, for HPLC, >=99.8%, contains 0.5-1.0% ethanol as stabilizer; Chloroform-[d]; Chloroform, 99.9%, Extra Dry over Molecular Sieve, Stabilized, AcroSeal(R); Chloroform, 99.9%, for residue analysis, ECD tested for pesticide anal., residue <0.0005%; trichloro- methane; Chloroform solution; methane trichloride; tris(chloranyl)methane; CCl3H; HCCl3; Chloroform, for HPLC; Trichloromethane, 9CI; Chloroform with amylene; Chloroform, ethanol-free; chloroformium pro narcosi; WLN: GYGG; Aqualine™ Matrix K; EC 200-663-8; 4-01-00-00042 (Beilstein Handbook Reference); CHEMBL44618; Chloroform, Environmental Grade; Chloroform, p.a., 99.8%; GTPL2503; Chloroform - Reagent Grade ACS; DTXSID1020306; R 20 (VAN); Chloroform with Amylene HPLC grade; Chloroform, Spectrophotometric Grade; NSC77361; ZINC8214524; Chloroform (stabilized with ethanol); Tox21_111024; Tox21_202494; 8296AF; Chloroform, for HPLC, >=99.5%; Chloroform, for HPLC, >=99.8%; NSC-77361; AKOS000269026; Chloroform 100 microg/mL in Methanol; DB11387; MCULE-5607930311; UN 1888; Chloroform 5000 microg/mL in Methanol; Chloroform, purification grade, >=99%; NCGC00090794-02; NCGC00260043-01; F 20; Trichloromethane 10 microg/mL in Methanol; Chloroform, JIS special grade, >=99.0%; Trichloromethane 100 microg/mL in Methanol; C0819; FT-0623661; Trichloromethane 5000 microg/mL in Methanol; Y1314; C13827; Chloroform, HPLC grade stabilized with ethanol; Chloroform (stabilized with 2-Methyl-2-butene); Chloroform, SAJ super special grade, >=99.0%; A835850; L023971; Q172275; BRD-K88785477-001-01-8; Chloroform, for HPLC, >=99.8%, amylene stabilized; Chloroform, 99.8%, ACS Reagent stabilized with Ethanol; Chloroform, technical, amylene stabilized, >=99% (GC); F0001-1775; Chloroform, technical grade, 95%, contains 50 ppm Amylene; Chloroform, 99.8%, for spectroscopy, stabilized with ethanol; Chloroform, anhydrous, contains amylenes as stabilizer, >=99%; Chloroform, for HPLC, >=99.8% (chloroform + ethanol, GC); Chloroform, for residue analysis, suitable for 5000 per JIS; Chloroform solution, 200 mug/mL in methanol, analytical standard; Chloroform stabilized with 50-200 ppm Amylene ACS Reagent Grade; Chloroform, >=99%, PCR Reagent, contains amylenes as stabilizer; Chloroform, ACS reagent, >=99.8%, contains amylenes as stabilizer; Chloroform, AR, contains 1-2% ethanol as stabilizer, >=99.5%; Chloroform, AR, contains 100 ppm amylene as stabilizer, >=99.5%; Chloroform, contains 100-200 ppm amylenes as stabilizer, >=99.5%; Chloroform, contains amylenes as stabilizer, ACS reagent, >=99.8%; Chloroform, contains ethanol as stabilizer, ACS reagent, >=99.8%; Chloroform, LR, contains 100 ppm amylene as stabilizer, >=99%; Chloroform, p.a., ACS reagent, 99.8%, contains 0.005% Amylene; Chloroform, SAJ first grade, >=99.0%, contains 0.4-0.8% ethanol; Chloroform, UV HPLC spectroscopic, 99.9%, contains 50 ppm Amylene; Chloroform solution, certified reference material, 5000 mug/mL in methanol; Chloroform, ACS reagent, Reag. Ph. Eur., contains ethanol as stabilizer; Chloroform, anhydrous, >=99%, contains 0.5-1.0% ethanol as stabilizer; Chloroform, Pharmaceutical Secondary Standard; Certified Reference Material; Chloroform, puriss. p.a., reag. ISO, reag. Ph. Eur., 99.0-99.4% (GC); Chloroform, suitable for 300 per JIS, >=99.0%, for residue analysis; Chloroform, UV HPLC spectroscopic, 99.0%, contains 0.6-1.0% Ethanol; Chloroform solution, NIST standard reference material, 1% in acetone-d6 (99.9 atom % D); Chloroform solution, NMR reference standard, 1% in acetone-d6 (99.9 atom % D); Chloroform solution, NMR reference standard, 3% in acetone-d6 (99.9 atom % D); Chloroform, ACS reagent, >=99.8%, contains 0.5-1.0% ethanol as stabilizer; Chloroform, ACS spectrophotometric grade, >=99.8%, contains amylenes as stabilizer; Chloroform, biotech. grade, >=99.8%, contains 0.5-1.0% ethanol as stabilizer; Chloroform, p.a., ACS reagent, reag. ISO, 99.8%, contains 50 ppm Amylene; Chloroform, puriss. p.a., ACS reagent, >=99.8% (chloroform + ethanol, GC); Chloroform, ReagentPlus(R), >=99.8%, contains 0.5-1.0% ethanol as stabilizer; Residual Solvent Class 2 - Chloroform, United States Pharmacopeia (USP) Reference Standard; 8013-54-5; Chloroform solution, NMR reference standard, 0.3% in acetone-d6 (99.9 atom % D), NMR tube size 3 mm x 8 in.; Chloroform solution, NMR reference standard, 0.3% in acetone-d6 (99.9 atom % D), NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 1% in acetone-d6 (99,9 atom % D), TMS 0.1 %, NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 1% in acetone-d6 (99.9 atom % D), NMR tube size 10 mm x 8 in.; Chloroform solution, NMR reference standard, 1% in acetone-d6 (99.9 atom % D), NMR tube size 3 mm x 8 in.; Chloroform solution, NMR reference standard, 1% in acetone-d6 (99.9 atom % D), NMR tube size 4 mm x 8 in.; Chloroform solution, NMR reference standard, 1% in acetone-d6 (99.9 atom % D), NMR tube size 8 mm x 8 in.; Chloroform solution, NMR reference standard, 10% in acetone-d6 (99.9 atom % D), NMR tube size 3 mm x 8 in.; Chloroform solution, NMR reference standard, 2% in acetone-d6 (99.9 atom % D), NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 2% in chloroform-d (99.8 atom % D), NMR tube size 3 mm x 8 in.; Chloroform solution, NMR reference standard, 2% in chloroform-d (99.8 atom % D), NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 20% in acetone-d6 (99.9 atom % D), NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 3% in acetone-d6 (99.9 atom % D), TMS 0.2 %, NMR tube size 3 mm x 8 in.; Chloroform solution, NMR reference standard, 3% in acetone-d6 (99.9 atom % D), TMS 0.2 %, NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 5% in acetone-d6 (99.9 atom % D), chromium(III) acetylacetonate 0.2 %; Chloroform solution, NMR reference standard, 5% in acetone-d6 (99.9 atom % D), chromium(III) acetylacetonate 2 %, NMR tube size 10 mm x 8 in.; Chloroform solution, NMR reference standard, 5% in acetone-d6 (99.9 atom % D), NMR tube size 3 mm x 8 in.; Chloroform solution, NMR reference standard, 5% in acetone-d6 (99.9 atom % D), NMR tube size 5 mm x 8 in.; Chloroform solution, NMR reference standard, 50% in acetone-d6 (99.9 atom % D), chromium(III) acetylacetonate 0.2 %; Chloroform solution, NMR reference standard, 50% in acetone-d6 (99.9 atom % D), chromium(III) acetylacetonate 0.2 %, NMR tube size 10 mm x 8 in.; Chloroform, ACS spectrophotometric grade, >=99.8%, contains 0.5-1.0% ethanol as stabilizer; Chloroform, contains ethanol as stabilizer, meets analytical specification of DAB9, BP, 99-99.4% (GC); Chloroform, HPLC Plus, for HPLC, GC, and residue analysis, >=99.9%, contains 0.5-1.0% ethanol as stabilizer; Chloroform, HPLC Plus, for HPLC, GC, and residue analysis, >=99.9%, contains amylenes as stabilizer; Chloroform/isoamyl alcohol 24:1(v/v), for molecular biology, DNAse, RNAse and Protease free; Residual Solvent - Chloroform, Pharmaceutical Secondary Standard; Certified Reference Material
Search Links to Other Online Resources (may not be available):
- Google Structure Search
- EPA CompTox Database
- Chemical Entities of Biological Interest (ChEBI)
- NIH ChemIDplus - A TOXNET DATABASE
- The Human Metabolome Database (HMDB)
- OECD eChemPortal
- Google Scholar
+==================================================+
|| Molecular Calculation ||
+==================================================+
Id = 63004
NWOutput = Link to NWChem Output (download)
Datafiles:
density.cube-518811-2021-9-1-12:37:2 (download)
homo-restricted.cube-518811-2021-9-1-12:37:2 (download)
lumo-restricted.cube-518811-2021-9-1-12:37:2 (download)
image_resset: api/image_reset/63004
Calculation performed by Eric Bylaska - arrow15.emsl.pnl.gov
Numbers of cpus used for calculation = 32
Calculation walltime = 637.300000 seconds (0 days 0 hours 10 minutes 37 seconds)
+----------------+
| Energetic Data |
+----------------+
Id = 63004
iupac = chloroform cation
mformula = C1Cl3
inchi = InChI=1S/CCl3/c2-1(3)4
inchikey = ZBZJXHCVGLJWFG-UHFFFAOYSA-N
esmiles = Cl[C](Cl)Cl theory{pspw} xc{pbe} basis{100.0 Ry} solvation_type{None} ^{1}
calculation_type = ov
theory = pspw
xc = pbe
basis = 100.0 Ry
charge,mult = 1 1
energy = -50.871588 Hartrees
enthalpy correct.= 0.013601 Hartrees
entropy = 71.177 cal/mol-K
solvation energy = 0.000 kcal/mol solvation_type = None
Sitkoff cavity dispersion = 1.943 kcal/mol
Honig cavity dispersion = 5.415 kcal/mol
ASA solvent accesible surface area = 216.620 Angstrom2
ASA solvent accesible volume = 227.356 Angstrom3
+-----------------+
| Structural Data |
+-----------------+
JSmol: an open-source HTML5 viewer for chemical structures in 3D
Number of Atoms = 4
Units are Angstrom for bonds and degrees for angles
Type I J K L M Value
----------- ----- ----- ----- ----- ----- ----------
1 Stretch Cl1 C2 1.64797
2 Stretch C2 Cl3 1.64794
3 Stretch C2 Cl4 1.64779
4 Bend Cl1 C2 Cl3 120.03459
5 Bend Cl1 C2 Cl4 119.97239
6 Bend Cl3 C2 Cl4 119.99298
+-----------------+
| Eigenvalue Data |
+-----------------+
Id = 63004
iupac = chloroform cation
mformula = C1Cl3
InChI = InChI=1S/CCl3/c2-1(3)4
smiles = Cl[C](Cl)Cl
esmiles = Cl[C](Cl)Cl theory{pspw} xc{pbe} basis{100.0 Ry} solvation_type{None} ^{1}
theory = pspw
xc = pbe
basis = 100.0 Ry
charge = 1
mult = 1
solvation_type = None
twirl webpage = TwirlMol Link
image webpage = GIF Image Link
Eigevalue Spectra
---------- 0.97 eV
---- ----
----------
---- ----
----------
---------- LUMO= -7.31 eV
HOMO= -10.47 eV ++++++++++
++ ++ ++ +
++++++++++
++++ ++++
++++++++++
++++ ++++
-27.90 eV ++++++++++
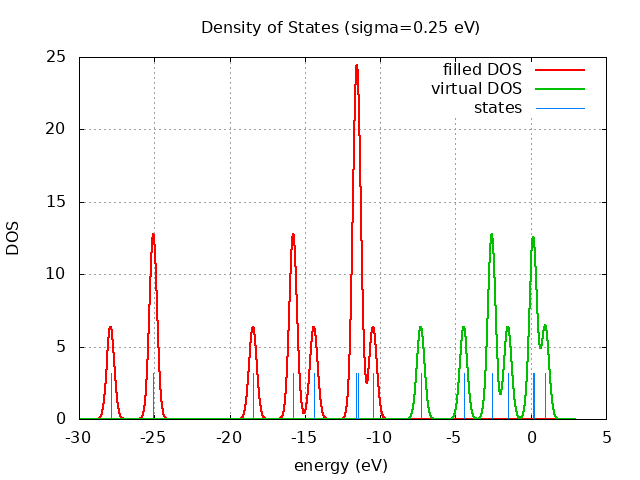
spin eig occ ---------------------------- restricted -10.47 2.00 restricted -11.48 2.00 restricted -11.48 2.00 restricted -11.62 2.00 restricted -11.63 2.00 restricted -14.41 2.00 restricted -15.78 2.00 restricted -15.78 2.00 restricted -18.45 2.00 restricted -25.07 2.00 restricted -25.07 2.00 restricted -27.90 2.00 restricted 0.97 0.00 restricted 0.21 0.00 restricted 0.12 0.00 restricted -1.52 0.00 restricted -2.60 0.00 restricted -2.60 0.00 restricted -4.46 0.00 restricted -7.31 0.00
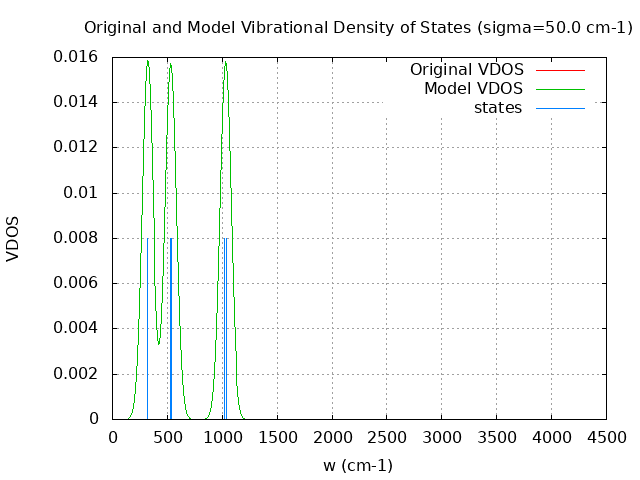
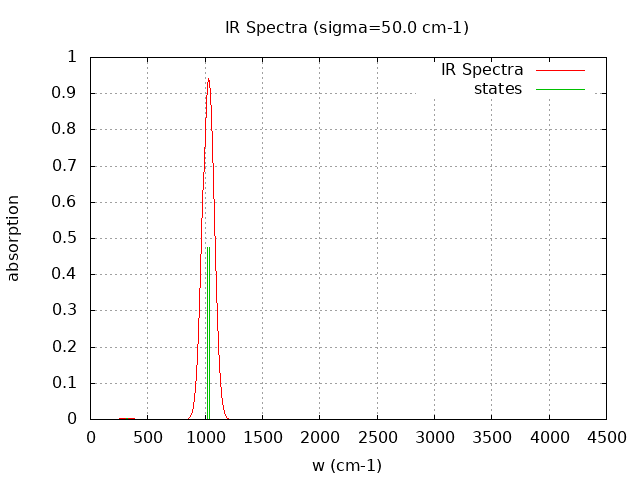
+----------------------------------------+ | Vibrational Density of States Analysis | +----------------------------------------+ Total number of frequencies = 12 Total number of negative frequencies = 0 Number of lowest frequencies = 2 (frequency threshold = 500 ) Exact dos norm = 6.000 Generating vibrational DOS Generating model vibrational DOS to have a proper norm 10.00 6.00 2.00 6.00 50.00 6.00 2.00 6.00 100.00 6.00 2.00 6.00 Generating IR Spectra Writing vibrational density of states (DOS) to vdos.dat Writing model vibrational density of states (DOS_FIXED) to vdos-model.dat Writing IR spectra to irdos.dat Temperature= 298.15 zero-point correction to energy = 5.375 kcal/mol ( 0.008566) vibrational contribution to enthalpy correction = 6.166 kcal/mol ( 0.009826) vibrational contribution to Entropy = 3.956 cal/mol-k hindered rotor enthalpy correction = 0.000 kcal/mol ( 0.000000) hindered rotor entropy correction = 0.000 cal/mol-k
DOS sigma = 10.000000
- vibrational DOS enthalpy correction = 0.009827 kcal/mol ( 6.167 kcal/mol)
- model vibrational DOS enthalpy correction = 0.009827 kcal/mol ( 6.167 kcal/mol)
- vibrational DOS Entropy = 0.000006 ( 3.959 cal/mol-k)
- model vibrational DOS Entropy = 0.000006 ( 3.960 cal/mol-k)
- original gas Energy = -50.871588 (-31922.403 kcal/mol)
- original gas Enthalpy = -50.857987 (-31913.869 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -50.857987 (-31913.868 kcal/mol, delta= 0.000)
- model DOS gas Enthalpy = -50.857986 (-31913.868 kcal/mol, delta= 0.001)
- original gas Entropy = 0.000113 ( 71.177 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000113 ( 71.180 cal/mol-k,delta= 0.003)
- model DOS gas Entropy = 0.000113 ( 71.181 cal/mol-k,delta= 0.004)
- original gas Free Energy = -50.891806 (-31935.090 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -50.891807 (-31935.091 kcal/mol, delta= -0.001)
- model DOS gas Free Energy = -50.891806 (-31935.090 kcal/mol, delta= -0.000)
- original sol Free Energy = -50.891806 (-31935.090 kcal/mol)
- unadjusted DOS sol Free Energy = -50.891807 (-31935.091 kcal/mol)
- model DOS sol Free Energy = -50.891806 (-31935.090 kcal/mol)
DOS sigma = 50.000000
- vibrational DOS enthalpy correction = 0.009840 kcal/mol ( 6.175 kcal/mol)
- model vibrational DOS enthalpy correction = 0.009840 kcal/mol ( 6.175 kcal/mol)
- vibrational DOS Entropy = 0.000006 ( 4.038 cal/mol-k)
- model vibrational DOS Entropy = 0.000006 ( 4.038 cal/mol-k)
- original gas Energy = -50.871588 (-31922.403 kcal/mol)
- original gas Enthalpy = -50.857987 (-31913.869 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -50.857974 (-31913.860 kcal/mol, delta= 0.008)
- model DOS gas Enthalpy = -50.857974 (-31913.860 kcal/mol, delta= 0.008)
- original gas Entropy = 0.000113 ( 71.177 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000114 ( 71.259 cal/mol-k,delta= 0.082)
- model DOS gas Entropy = 0.000114 ( 71.259 cal/mol-k,delta= 0.082)
- original gas Free Energy = -50.891806 (-31935.090 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -50.891831 (-31935.106 kcal/mol, delta= -0.016)
- model DOS gas Free Energy = -50.891831 (-31935.106 kcal/mol, delta= -0.016)
- original sol Free Energy = -50.891806 (-31935.090 kcal/mol)
- unadjusted DOS sol Free Energy = -50.891831 (-31935.106 kcal/mol)
- model DOS sol Free Energy = -50.891831 (-31935.106 kcal/mol)
DOS sigma = 100.000000
- vibrational DOS enthalpy correction = 0.009879 kcal/mol ( 6.199 kcal/mol)
- model vibrational DOS enthalpy correction = 0.009880 kcal/mol ( 6.200 kcal/mol)
- vibrational DOS Entropy = 0.000007 ( 4.321 cal/mol-k)
- model vibrational DOS Entropy = 0.000007 ( 4.323 cal/mol-k)
- original gas Energy = -50.871588 (-31922.403 kcal/mol)
- original gas Enthalpy = -50.857987 (-31913.869 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -50.857935 (-31913.836 kcal/mol, delta= 0.033)
- model DOS gas Enthalpy = -50.857933 (-31913.835 kcal/mol, delta= 0.034)
- original gas Entropy = 0.000113 ( 71.177 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000114 ( 71.541 cal/mol-k,delta= 0.364)
- model DOS gas Entropy = 0.000114 ( 71.544 cal/mol-k,delta= 0.367)
- original gas Free Energy = -50.891806 (-31935.090 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -50.891927 (-31935.166 kcal/mol, delta= -0.076)
- model DOS gas Free Energy = -50.891926 (-31935.165 kcal/mol, delta= -0.075)
- original sol Free Energy = -50.891806 (-31935.090 kcal/mol)
- unadjusted DOS sol Free Energy = -50.891927 (-31935.166 kcal/mol)
- model DOS sol Free Energy = -50.891926 (-31935.165 kcal/mol)
Normal Mode Frequency (cm-1) IR Intensity (arbitrary)
----------- ---------------- ------------------------
1 0.000 0.013
2 0.000 0.011
3 0.000 0.091
4 0.000 0.076
5 0.000 0.005
6 0.000 0.068
7 315.140 0.240
8 322.680 0.226
9 525.130 0.124
10 541.970 0.009
11 1021.920 59.578
12 1034.910 59.559
No Hindered Rotor Data
+-------------------------------------+
| Reactions Contained in the Database |
+-------------------------------------+
Reactions Containing INCHIKEY = ZBZJXHCVGLJWFG-UHFFFAOYSA-N
Reactionid Erxn(gas) Hrxn(gas) Grxn(gas) delta_Solv Grxn(aq) ReactionType Reaction
20975 -34.335 -35.010 -42.055 -71.900 -15.355 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe} + SHE xc{pbe} --> [C](Cl)(Cl)Cl ^{-1} xc{pbe} + [Cl-] xc{pbe}"
20974 -34.335 -35.010 -42.055 -71.900 -15.355 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe} + SHE xc{pbe} --> [C](Cl)(Cl)Cl ^{-1} xc{pbe} + [Cl-] xc{pbe}"
20932 -9.261 -9.367 -2.942 26.220 23.278 A + B --> AB "Cl[C](Cl)Cl + [Cl-] --> carbon tetrachloride ^{-1}"
20918 -357.268 -350.155 -341.630 304.909 -36.721 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{pbe} + [H+] xc{pbe} --> C(Cl)(Cl)Cl xc{pbe}"
20864 33.171 33.016 25.391 -32.406 -7.015 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
20863 33.171 33.016 25.391 -32.406 -7.015 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
20773 -406.430 -400.186 -392.506 0.000 -293.906 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} + SHE theory{pspw4} + [H+] theory{pspw4} --> C(Cl)(Cl)Cl theory{pspw4}"
20722 -40.576 -41.388 -48.632 -80.135 -30.168 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [Cl-] xc{b3lyp}"
20721 -40.576 -41.388 -48.632 -80.135 -30.168 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [Cl-] xc{b3lyp}"
20632 -412.681 -406.160 -398.450 256.094 -43.756 A + B --> AB "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl xc{b3lyp}"
20629 9.261 9.367 2.942 -26.220 -23.278 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
20628 9.261 9.367 2.942 -26.220 -23.278 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
20575 -24.379 -25.774 -29.866 0.000 -29.866 AB + C --> AC + B "[K] theory{pspw} + ClC(Cl)(Cl)Cl theory{pspw} --> Cl[K] theory{pspw} + Cl[C](Cl)Cl theory{pspw}"
20515 -362.844 -355.405 -346.876 310.009 -36.867 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl xc{b3lyp}"
20443 -364.926 -357.734 -349.389 0.000 -349.389 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} + [H+] theory{pspw4} --> C(Cl)(Cl)Cl theory{pspw4}"
19838 -102.150 -102.030 -92.756 41.423 -51.334 A + B --> AB "[C](Cl)(Cl)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> C(Cl)(Cl)(Cl)O ^{-1} xc{m06-2x}"
18752 159.015 152.796 145.500 -193.415 -47.915 AB --> A + B "ClC(Cl)Cl ^{1} mult{2} --> Cl[C](Cl)Cl mult{2} + [H] ^{1}"
18751 159.015 152.796 145.500 -193.415 -47.915 AB --> A + B "ClC(Cl)Cl ^{1} mult{2} --> Cl[C](Cl)Cl mult{2} + [H] ^{1}"
18231 -155.975 -156.251 -147.939 131.441 -16.498 A + B --> AB "[Br-] xc{pbe} + [C](Cl)(Cl)Cl ^{+1} xc{pbe} --> BrC(Cl)(Cl)Cl xc{pbe}"
17208 -21.902 -22.965 -31.438 -81.432 -14.271 AB --> A + B "Cl[C](Cl)Cl mult{2} + [SHE] --> Cl[C]Cl + [Cl] ^{-1}"
17207 -21.902 -22.965 -31.438 -81.432 -14.271 AB --> A + B "Cl[C](Cl)Cl mult{2} + [SHE] --> Cl[C]Cl + [Cl] ^{-1}"
15552 -410.114 -403.595 -395.886 256.054 -41.232 A + B --> AB "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl xc{b3lyp}"
15541 -357.269 -350.156 -341.629 309.969 -31.660 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{pbe} + [H+] xc{pbe} --> C(Cl)(Cl)Cl xc{pbe}"
15531 -360.277 -352.840 -344.312 309.969 -34.343 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl xc{b3lyp}"
15447 -405.161 -398.941 -391.228 256.185 -36.444 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe} + SHE xc{pbe} + [H+] xc{pbe} --> C(Cl)(Cl)Cl xc{pbe}"
14745 -100.836 -100.188 -93.968 48.274 -45.694 A + B --> AB "[C](Cl)(Cl)Cl theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> [C-](Cl)(Cl)(Cl)O theory{ccsd(t)}"
12975 -18.084 -18.616 -27.574 0.000 71.026 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{pspw4} + [SHE] theory{pspw4} --> [C](Cl)(Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
12974 -18.084 -18.616 -27.574 0.000 71.026 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{pspw4} + [SHE] theory{pspw4} --> [C](Cl)(Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
12879 -23.232 -24.652 -28.334 0.000 -28.334 AB + C --> AC + B "[K] theory{pspw4} + ClC(Cl)(Cl)Cl theory{pspw4} --> Cl[K] theory{pspw4} + Cl[C](Cl)Cl theory{pspw4}"
12804 -363.517 -355.914 -347.615 310.836 -36.779 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)(Cl)Cl xc{m06-2x}"
12677 -413.205 -406.519 -398.775 255.974 -44.202 A + B --> AB "[C](Cl)(Cl)Cl xc{m06-2x} + SHE xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)(Cl)Cl xc{m06-2x}"
12451 -101.397 -101.199 -93.337 50.638 -42.699 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe0} + [OH-] xc{pbe0} --> C(Cl)(Cl)(Cl)O ^{-1} xc{pbe0}"
8010 199.671 199.526 191.872 -236.037 54.435 AB --> A + B "Cl[C](Cl)Cl ^{-1} + [SHE] --> Cl[C]Cl + [Cl] ^{-2} mult{2}"
8009 199.671 199.526 191.872 -236.037 54.435 AB --> A + B "Cl[C](Cl)Cl ^{-1} + [SHE] --> Cl[C]Cl + [Cl] ^{-2} mult{2}"
7969 371.510 369.871 360.737 -157.131 203.606 AB --> A + B "Cl[C](Cl)Cl mult{2} --> Cl[C]Cl ^{-1} mult{2} + [Cl] ^{1}"
7968 371.510 369.871 360.737 -157.131 203.606 AB --> A + B "Cl[C](Cl)Cl mult{2} --> Cl[C]Cl ^{-1} mult{2} + [Cl] ^{1}"
7742 -9.252 -9.323 -2.784 20.459 17.675 A + B --> AB "Cl[C](Cl)Cl + [Cl-] --> carbon tetrachloride ^{-1}"
7239 -371.374 -363.842 -355.609 0.000 -355.609 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} + [H+] theory{pspw4} xc{pbe0} --> C(Cl)(Cl)Cl theory{pspw4} xc{pbe0}"
7238 31.347 31.099 23.018 0.000 23.018 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} --> [C](Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7237 31.347 31.099 23.018 0.000 23.018 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} --> [C](Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7236 -30.195 -31.209 -38.804 0.000 59.796 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7235 -30.195 -31.209 -38.804 0.000 59.796 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7234 7.366 7.336 0.238 0.000 0.238 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} --> [C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7233 7.366 7.336 0.238 0.000 0.238 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} --> [C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7232 3.554 2.605 -5.868 0.000 92.732 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)Cl mult{3} theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7231 3.554 2.605 -5.868 0.000 92.732 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)Cl mult{3} theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7228 -152.812 -146.592 -139.296 193.415 54.119 A + B --> AB "[C](Cl)(Cl)Cl theory{ccsd(t)} + [H+] theory{ccsd(t)} --> C(Cl)(Cl)Cl ^{+1} theory{ccsd(t)}"
7216 -100.836 -100.188 -93.967 48.354 -45.613 A + B --> AB "[C](Cl)(Cl)Cl theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> [C-](Cl)(Cl)(Cl)O theory{ccsd(t)}"
7213 -40.585 -41.433 -48.790 -74.375 -24.564 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [Cl-] xc{b3lyp}"
7212 -40.585 -41.433 -48.790 -74.375 -24.564 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [Cl-] xc{b3lyp}"
7211 9.252 9.323 2.784 -20.459 -17.675 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7210 9.252 9.323 2.784 -20.459 -17.675 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7209 -35.528 -36.375 -43.732 -74.375 -19.507 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7208 -35.528 -36.375 -43.732 -74.375 -19.507 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7207 9.199 9.270 2.732 -20.459 -17.727 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl theory{ccsd(t)} --> [C](Cl)(Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7206 9.199 9.270 2.732 -20.459 -17.727 AB --> A + B "[C-](Cl)(Cl)(Cl)Cl theory{ccsd(t)} --> [C](Cl)(Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7190 30.204 30.059 22.404 -27.517 -5.113 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} --> [C](Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7189 30.204 30.059 22.404 -27.517 -5.113 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} --> [C](Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7188 -34.254 -35.151 -42.374 -74.412 -18.186 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe0} + SHE xc{pbe0} --> [C](Cl)(Cl)Cl ^{-1} xc{pbe0} + [Cl-] xc{pbe0}"
7187 -34.254 -35.151 -42.374 -74.412 -18.186 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe0} + SHE xc{pbe0} --> [C](Cl)(Cl)Cl ^{-1} xc{pbe0} + [Cl-] xc{pbe0}"
7186 9.322 9.356 2.750 -19.462 -16.711 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe0} --> [C](Cl)(Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
7185 9.322 9.356 2.750 -19.462 -16.711 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe0} --> [C](Cl)(Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
7184 -93.655 -92.290 -83.267 0.000 -83.267 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} + [OH-] theory{pspw4} --> C(Cl)(Cl)(Cl)O ^{-1} theory{pspw4}"
7181 -102.150 -102.031 -92.757 47.273 -45.484 A + B --> AB "[C](Cl)(Cl)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> C(Cl)(Cl)(Cl)O ^{-1} xc{m06-2x}"
7180 4.597 3.744 -4.666 0.000 93.934 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} + SHE theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
7179 4.597 3.744 -4.666 0.000 93.934 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} + SHE theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
7174 -58.258 -55.234 -45.068 0.000 -45.068 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + [OH-] theory{pspw4} xc{pbe0} --> C(Cl)(Cl)(Cl)O ^{-1} theory{pspw4} xc{pbe0}"
7173 -101.397 -101.199 -93.337 50.698 -42.639 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe0} + [OH-] xc{pbe0} --> C(Cl)(Cl)(Cl)O ^{-1} xc{pbe0}"
7172 -103.062 -102.691 -95.247 49.601 -45.646 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe} + [OH-] xc{pbe} --> C(Cl)(Cl)(Cl)O ^{-1} xc{pbe}"
7164 -34.334 -35.010 -42.057 -76.959 -20.416 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe} + SHE xc{pbe} --> [C](Cl)(Cl)Cl ^{-1} xc{pbe} + [Cl-] xc{pbe}"
7163 -34.334 -35.010 -42.057 -76.959 -20.416 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe} + SHE xc{pbe} --> [C](Cl)(Cl)Cl ^{-1} xc{pbe} + [Cl-] xc{pbe}"
7162 13.558 13.775 7.543 -23.175 -15.632 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)(Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
7161 13.558 13.775 7.543 -23.175 -15.632 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)(Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
7141 33.003 32.766 24.802 -27.199 -2.397 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
7140 33.003 32.766 24.802 -27.199 -2.397 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
7139 32.322 32.121 24.235 -27.200 -2.965 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
7138 32.322 32.121 24.235 -27.200 -2.965 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
7137 33.170 33.015 25.392 -27.347 -1.954 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
7136 33.170 33.015 25.392 -27.347 -1.954 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
7129 -155.333 -149.028 -141.420 0.000 -141.420 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + [H+] theory{pspw4} xc{pbe0} --> C(Cl)(Cl)Cl ^{+1} theory{pspw4} xc{pbe0}"
7128 -408.935 -402.388 -394.651 0.000 -296.051 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} + [H+] theory{pspw4} xc{pbe0} --> C(Cl)(Cl)Cl theory{pspw4} xc{pbe0}"
7127 -18.764 -19.430 -28.508 0.000 70.092 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7126 -18.764 -19.430 -28.508 0.000 70.092 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7125 -37.878 -38.784 -46.172 -74.731 -22.303 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{m06-2x} + SHE xc{m06-2x} --> [C](Cl)(Cl)Cl ^{-1} xc{m06-2x} + [Cl-] xc{m06-2x}"
7124 -37.878 -38.784 -46.172 -74.731 -22.303 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{m06-2x} + SHE xc{m06-2x} --> [C](Cl)(Cl)Cl ^{-1} xc{m06-2x} + [Cl-] xc{m06-2x}"
7123 11.810 11.821 4.989 -19.869 -14.880 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{m06-2x} --> [C](Cl)(Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
7122 11.810 11.821 4.989 -19.869 -14.880 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{m06-2x} --> [C](Cl)(Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
7121 -163.373 -157.272 -149.682 0.000 -149.682 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} + [H+] theory{pspw4} --> C(Cl)(Cl)Cl ^{+1} theory{pspw4}"
7120 -367.603 -360.179 -351.652 309.939 -41.713 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} + [H+] theory{ccsd(t)} --> C(Cl)(Cl)Cl theory{ccsd(t)}"
7119 5.281 4.430 -3.948 -80.119 14.533 AB --> A + B "[C](Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7118 5.281 4.430 -3.948 -80.119 14.533 AB --> A + B "[C](Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7117 1.670 0.749 -7.693 -80.697 10.210 AB --> A + B "[C](Cl)(Cl)Cl xc{m06-2x} + SHE xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
7116 1.670 0.749 -7.693 -80.697 10.210 AB --> A + B "[C](Cl)(Cl)Cl xc{m06-2x} + SHE xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
7115 -412.330 -405.824 -398.116 256.024 -43.492 A + B --> AB "[C](Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} + [H+] theory{ccsd(t)} --> C(Cl)(Cl)Cl theory{ccsd(t)}"
7114 -12.411 -13.067 -22.046 -79.556 -3.001 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)(Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7113 -12.411 -13.067 -22.046 -79.556 -3.001 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)(Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7111 -147.420 -142.661 -134.912 191.799 56.887 A + B --> AB "[C](Cl)(Cl)Cl xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)(Cl)Cl ^{+1} xc{m06-2x}"
7110 -153.111 -148.255 -140.517 192.070 51.553 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe0} + [H+] xc{pbe0} --> C(Cl)(Cl)Cl ^{+1} xc{pbe0}"
7109 -161.346 -155.986 -148.063 192.532 44.469 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe} + [H+] xc{pbe} --> C(Cl)(Cl)Cl ^{+1} xc{pbe}"
7108 -159.015 -152.796 -145.500 193.415 47.915 A + B --> AB "[C](Cl)(Cl)Cl xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl ^{+1} xc{b3lyp}"
7104 -364.925 -357.742 -349.368 0.000 -349.368 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} + [H+] theory{pspw4} --> C(Cl)(Cl)Cl theory{pspw4}"
7103 -363.512 -355.904 -347.605 310.845 -36.760 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)(Cl)Cl xc{m06-2x}"
7102 -366.655 -359.204 -350.860 310.974 -39.886 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{pbe0} + [H+] xc{pbe0} --> C(Cl)(Cl)Cl xc{pbe0}"
7101 -360.340 -353.230 -344.706 309.989 -34.718 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{pbe} + [H+] xc{pbe} --> C(Cl)(Cl)Cl xc{pbe}"
7100 -362.843 -355.419 -346.892 309.939 -36.953 A + B --> AB "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl xc{b3lyp}"
7099 31.830 31.599 23.703 0.000 23.703 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} --> [C](Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
7098 31.830 31.599 23.703 0.000 23.703 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} --> [C](Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
7097 27.935 27.790 20.136 -27.517 -7.381 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} --> [C](Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7096 27.935 27.790 20.136 -27.517 -7.381 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} --> [C](Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7092 -34.982 -35.303 -42.866 0.000 55.734 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} theory{pspw4} + SHE theory{pspw4} --> [C](Cl)(Cl)Cl ^{-1} theory{pspw4} + [Cl-] theory{pspw4}"
7091 -34.982 -35.303 -42.866 0.000 55.734 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} theory{pspw4} + SHE theory{pspw4} --> [C](Cl)(Cl)Cl ^{-1} theory{pspw4} + [Cl-] theory{pspw4}"
7076 -46.923 -47.562 -55.903 -85.108 -42.412 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [Cl-] xc{b3lyp}"
7075 -46.923 -47.562 -55.903 -85.108 -42.412 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)(Cl)Cl ^{-1} xc{b3lyp} + [Cl-] xc{b3lyp}"
7074 6.524 7.150 0.252 0.000 0.252 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} theory{pspw4} --> [C](Cl)(Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
7073 6.524 7.150 0.252 0.000 0.252 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} theory{pspw4} --> [C](Cl)(Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
7072 2.914 3.193 -4.329 -31.193 -35.522 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7071 2.914 3.193 -4.329 -31.193 -35.522 AB --> A + B "C(Cl)(Cl)(Cl)Cl ^{-1} xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7070 3.231 2.327 -6.092 -80.708 11.800 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe0} + SHE xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
7069 3.231 2.327 -6.092 -80.708 11.800 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe0} + SHE xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
7068 4.846 4.051 -4.264 -79.950 14.386 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe} + SHE xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
7067 4.846 4.051 -4.264 -79.950 14.386 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe} + SHE xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
7066 -3.693 -4.543 -12.921 -80.119 5.559 AB --> A + B "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
7065 -3.693 -4.543 -12.921 -80.119 5.559 AB --> A + B "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
7064 -406.430 -400.195 -392.486 0.000 -293.886 A + B --> AB "[C](Cl)(Cl)Cl theory{pspw4} + SHE theory{pspw4} + [H+] theory{pspw4} --> C(Cl)(Cl)Cl theory{pspw4}"
7063 -413.200 -406.510 -398.765 255.983 -44.182 A + B --> AB "[C](Cl)(Cl)Cl xc{m06-2x} + SHE xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)(Cl)Cl xc{m06-2x}"
7062 -410.231 -403.711 -395.984 256.023 -41.361 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe0} + SHE xc{pbe0} + [H+] xc{pbe0} --> C(Cl)(Cl)Cl xc{pbe0}"
7061 -408.232 -402.015 -394.306 256.204 -39.502 A + B --> AB "[C](Cl)(Cl)Cl xc{pbe} + SHE xc{pbe} + [H+] xc{pbe} --> C(Cl)(Cl)Cl xc{pbe}"
7060 -412.680 -406.174 -398.466 256.024 -43.842 A + B --> AB "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)(Cl)Cl xc{b3lyp}"
7016 -37.707 -39.251 -42.869 -32.586 -75.455 AB + C --> AC + B "[K] + ClC(Cl)(Cl)Cl --> Cl[K] + Cl[C](Cl)Cl"
7003 -24.380 -25.791 -29.896 0.000 -29.896 AB + C --> AC + B "[K] theory{pspw} + ClC(Cl)(Cl)Cl theory{pspw} --> Cl[K] theory{pspw} + Cl[C](Cl)Cl theory{pspw}"
7002 -23.234 -24.668 -28.367 0.000 -28.367 AB + C --> AC + B "[K] theory{pspw4} + ClC(Cl)(Cl)Cl theory{pspw4} --> Cl[K] theory{pspw4} + Cl[C](Cl)Cl theory{pspw4}"
7001 -33.056 -34.600 -38.154 -25.593 -63.747 AB + C --> AC + B "[Na] + ClC(Cl)(Cl)Cl --> Cl[C](Cl)Cl + Cl[Na]"
5528 -111.834 -111.968 -111.389 4.279 -107.110 A + B --> AB "Cl[C](Cl)Cl ^{-1} + hydroxide ^{-1} --> OC(Cl)(Cl)Cl ^{-2}"
5526 -104.719 -104.071 -97.851 48.274 -49.577 A + B --> AB "Cl[C](Cl)Cl mult{2} + hydroxide ^{-1} --> OC(Cl)(Cl)Cl ^{-1} mult{2}"
5151 360.129 358.555 348.761 -152.440 196.321 AB --> A + B "C(Cl)(Cl)(Cl)Cl --> Cl[C](Cl)Cl ^{-1} + [Cl] ^{1}"
5150 360.129 358.555 348.761 -152.440 196.321 AB --> A + B "C(Cl)(Cl)(Cl)Cl --> Cl[C](Cl)Cl ^{-1} + [Cl] ^{1}"
2105 -15.786 -16.546 -25.636 -80.224 -7.261 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{m06-2x} + [SHE] xc{m06-2x} --> [C](Cl)(Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
2104 -15.786 -16.546 -25.636 -80.224 -7.261 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{m06-2x} + [SHE] xc{m06-2x} --> [C](Cl)(Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
2103 -25.475 -26.131 -35.106 -79.605 -16.110 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{b3lyp} + [SHE] xc{b3lyp} --> [C](Cl)(Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
2102 -16.924 -17.505 -26.435 -79.386 -7.221 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{pbe} + [SHE] xc{pbe} --> [C](Cl)(Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
2101 -16.924 -17.505 -26.435 -79.386 -7.221 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{pbe} + [SHE] xc{pbe} --> [C](Cl)(Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
2100 -18.042 -18.739 -27.776 -80.224 -9.400 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{pbe0} + [SHE] xc{pbe0} --> [C](Cl)(Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
2099 -18.042 -18.739 -27.776 -80.224 -9.400 AB --> A + B "C(Cl)(Cl)(Cl)Cl xc{pbe0} + [SHE] xc{pbe0} --> [C](Cl)(Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
2098 -18.085 -18.632 -27.606 0.000 70.994 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{pspw4} + [SHE] theory{pspw4} --> [C](Cl)(Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
2097 -18.085 -18.632 -27.606 0.000 70.994 AB --> A + B "C(Cl)(Cl)(Cl)Cl theory{pspw4} + [SHE] theory{pspw4} --> [C](Cl)(Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
1268 -2.914 -3.193 4.329 31.193 35.522 A + B --> AB "Cl[C](Cl)Cl + [Cl-] --> carbon tetrachloride ^{-1}"
820 25.475 26.131 35.106 79.605 16.110 A + B --> AB "[C](Cl)(Cl)Cl + [Cl-] --> carbon tetrachloride + SHE"
811 -25.475 -26.131 -35.106 -79.605 -16.110 AB --> A + B "carbon tetrachloride + [ SHE] --> [C](Cl)(Cl)Cl mult{2} + [Cl-]"
749 -3.693 -4.543 -12.921 -80.119 -93.041 AB --> A + B "Cl[C](Cl)Cl xyzdata{C -4.00022 0.58207 0.96997 | Cl -2.30229 0.97442 1.34210 | Cl -4.34205 -1.12742 1.31115 | Cl -4.38438 0.98070 -0.71795} --> [Cl] xyzdata{Cl 2.75521 1.83484 -0.24354} ^{-1} + Cl[C]Cl xyzdata{C 3.04352 -2.52273 -0.25387 | Cl 1.77592 -1.56644 -1.04794 | Cl 3.94514 -1.50287 0.88596} mult{3}"
All requests to Arrows were successful.
KEYWORDs -
reaction: :reaction
chinese_room: :chinese_room
molecule: :molecule
nmr: :nmr
predict: :predict
submitesmiles: :submitesmiles
nosubmitmissingesmiles
resubmitmissingesmiles
submitmachines: :submitmachines
useallentries
nomodelcorrect
eigenvalues: :eigenvalues
frequencies: :frequencies
nwoutput: :nwoutput
xyzfile: :xyzfile
alleigs: :alleigs
allfreqs: :allfreqs
reactionenumerate:
energytype:[erxn(gas) hrxn(gas) grxn(gas) delta_solvation grxn(aq)] :energytype
energytype:[kcal/mol kj/mol ev cm-1 ry hartree au] :energytype
tablereactions:
reaction: ... :reaction
reaction: ... :reaction
...
:tablereactions
tablemethods:
method: ... :method
method: ... :method
...
:tablemethods
:reactionenumerate
rotatebonds
xyzinput:
label: :label
xyzdata:
... xyz data ...
:xyzdata
:xyzinput
submitHf: :submitHf
nmrexp: :nmrexp
findreplace: old text | new text :findreplace
listnwjobs
fetchnwjob: :fetchnwjob
pushnwjob: :pushnwjob
printcsv: :printcsv
printeig: :printeig
printfreq: :printfreq
printxyz: :printxyz
printjobinfo: :printjobinfo
printnwout: :printnwout
badids: :badids
hup_string:
database:
table:
request_table:
listallesmiles
queuecheck
This software service and its documentation were developed at the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory, a multiprogram national laboratory, operated for the U.S. Department of Energy by Battelle under Contract Number DE-AC05-76RL01830. Support for this work was provided by the Department of Energy Office of Biological and Environmental Research, and Department of Defense environmental science and technology program (SERDP). THE SOFTWARE SERVICE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE SERVICE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE SERVICE.