 3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual
3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual Results from an EMSL Arrows Calculation
| EMSL Arrows is a revolutionary approach to materials and chemical simulations that uses NWChem and chemical computational databases to make materials and chemical modeling accessible via a broad spectrum of digital communications including posts to web APIs, social networks, and traditional email. |
Molecular modeling software has previously been extremely complex, making it prohibative to all but experts in the field, yet even experts can struggle to perform calculations. This service is designed to be used by experts and non-experts alike. Experts can carry out and keep track of large numbers of complex calculations with diverse levels of theories present in their workflows. Additionally, due to a streamlined and easy-to-use input, non-experts can carry out a wide variety of molecular modeling calculations previously not accessible to them.
The id(s) for emsiles = ClCCl theory{dft} xc{pbe0} basis{6-311++G(2d,2p)} solvation_type{COSMO} ^{0} are: 46454
Use id=% instead of esmiles to print other entries.
mformula = C1Cl2H2
iupac = dichloromethane
PubChem = 6344
PubChem LCSS = 6344
cas = 75-09-2
kegg = C02271 D02330
synonyms = DICHLOROMETHANE; Methylene chloride; Methylene dichloride; Methane, dichloro-; 75-09-2; Methylene bichloride; Methane dichloride; Solaesthin; Solmethine; Freon 30; Narkotil; Aerothene MM; Metylenu chlorek; Chlorure de methylene; Metaclen; Soleana VDA; CH2Cl2; Khladon 30; Dichlormethan; F 30 (chlorocarbon); Methylenum chloratum; RCRA waste number U080; NCI-C50102; R 30; R30 (refrigerant); Caswell No. 568; Methoklone; HCC 30; Salesthin; dichloro-methane; Methylene choride; Metylenu chlorek [Polish]; UNII-588X2YUY0A; CCRIS 392; HSDB 66; NSC 406122; UN 1593; Chlorure de methylene [French]; F 30; EINECS 200-838-9; UN1593; DICHLOROMETHANE, NF; RCRA waste no. U080; DICHLOROMETHANE, ACS; EPA Pesticide Chemical Code 042004; BRN 1730800; Dichloromethane, HPLC Grade; AI3-01773; METHYLENE CHLORIDE (DICHLOROMETHANE); CHEBI:15767; methylenechloride; YMWUJEATGCHHMB-UHFFFAOYSA-N; MFCD00000881; Chloride, Methylene; Bichloride, Methylene; Dichloride, Methylene; Methylene Chloride [USAN]; dichioromethane; dichlormetane; dichlormethane; dichloromeihane; dichlorometan; dichlorometane; dichloromethan; dichoromethane; dicloromethane; methylenchoride; metylenchloride; Methylenchlorid; Plastisolve; Solaesthie; Aerothene; Driverit; Narkotie; Nevolin; dichlor-methane; dichlorometliane; dichlorornethane; dicliloromethane; methylenchloride; methylenechlorid; methYIenechlorid; di-chloromethane; dichloro methane; dichloromethane-; methlyenechloride; Dichloromethane, suitable for 5000 per JIS, for residue analysis; methlene chloride; methyene chloride; methylen chloride; methylene chlorie; methylene cloride; metylene chloride; mehtylene chloride; methlyene chloride; methylene,chloride; methylene-chloride; Di-cle; dichloro -methane; dichloro- methane; methylenedichloride; Refrigerant 30; Distillex DS3; Dichloromethane, ACS reagent, >=99.5%, contains 40-150 ppm amylene as stabilizer; Dichloromethane (Methylenechloride); M-clean D; methyl ene chloride; Dichloromethane (Methylene Chloride); MeCl2; QMABHXaIh@; methylene di chloride; N,N-methylenechloride; AmbotzSOL-002; Methylene chloride ACS; Cl2CH2; H2CCl2; AC1L1MBL; Methylene chloride (NF); DSSTox_CID_868; Dichloromethane, anhydrous; ACMC-1BFR1; Dichloromethane, for HPLC; NCIMech_000221; AC1O5DP8; AC1Q3UP8; WLN: G1G; DSSTox_RID_75836; DSSTox_GSID_20868; 4-01-00-00035 (Beilstein Handbook Reference); Dichloromethane, >=99.9%; KSC237A3D; CHEMBL45967; Methylene Chloride HPLC grade; Methylene Chloride (Recovered); 588X2YUY0A; Dichloromethane, AR, >=99%; Freon 30, R-30, DCe; Dichloromethane (Peptide Grade); CTK1D7031; Dichloromethane Reagent Grade ACS; MolPort-001-757-423; UN 1593, MDC; Dichloromethane, LR, >=99.5%; Dichloromethane, purification grade; Dichloromethane, analytical standard; Dichloromethane, Environmental Grade; KS-00000W6J; Tox21_202526; ANW-42404; NSC406122; STL264204; Dichloromethane [UN1593] [Poison]; Methylene chloride-Methane, dichloro-; AKOS009031498; InChI=1/CH2Cl2/c2-1-3/h1H; Dichloromethane [UN1593] [Poison]; Dichloromethane, ACS reagent, 99.5%; LS-1655; MCULE-1114735075; NSC-406122; RL04864; RTR-037666; TRA0007256; CAS-75-09-2; Dichloromethane, for HPLC, >=99.7%; Dichloromethane GC, for residue analysis; Dichloromethane, Spectrophotometric Grade; NCGC00091504-01; NCGC00260075-01; AN-23837; KB-76510; SC-26935; Dichloromethane, suitable for PCB analysis; AB1007261; TR-037666; D0529; D3478; FT-0624716; M0629; Dichloromethane, for HPLC, >=99.8% (GC); Dichloromethane, SAJ first grade, >=99.0%; Dichloromethane, Selectophore(TM), >=99.5%; C02271; D02330; Dichloromethane, analytical standard, stabilized; Dichloromethane, JIS special grade, >=99.0%; Dideuteromethylenechloride; Methylene chloride-d2; 300062X; Dichloromethane, >71% in a non hazardous diluent; L023970; J-610006; Dichloromethane, 99%, stab. with ca. 50ppm amylene; I14-19640; Dichloromethane solution, contains 10 % (v/v) methanol; Dichloromethane, ACS, 99.5% min., stabilized 500ml; Dichloromethane, TLC high-purity grade, >=99.8% (GC); Dichloromethane HPLC, UV/IR, min. 99.9%, isocratic grade; Dichloromethane (Methylenechloride) 100 microg/mL in Methanol; Dichloromethane (Methylenechloride) 1000 ng/microl in Methanol; Dichloromethane, special, 99.9%, contains 40-60 ppm Amylene; Dichloromethane, for HPLC, >=99.8%, contains amylene as stabilizer; Dichloromethane, Selectophore(TM), >=99.5% (GC), inhibitor-free; Dichloromethane, suitable for 300 per JIS, for residue analysis; Dichloromethane, technical grade, 95%, contains 40-60 ppm Amylene; Methylene chloride, European Pharmacopoeia (EP) Reference Standard; Dichloromethane, puriss. p.a., ACS reagent, reag. ISO, >=99.9% (GC); Dichloromethane, UV HPLC spectroscopic, 99.9%, contains 40-60 ppm Amylene; Dichloromethane solution, 10 % (v/v) in methanol, 1 % (v/v) in ammonium hydroxide; Dichloromethane solution, certified reference material, 200 mug/mL in methanol; Dichloromethane solution, certified reference material, 5000 mug/mL in methanol; Dichloromethane, ACS reagent, >=99.5%, contains 50 ppm amylene as stabilizer; Dichloromethane, anhydrous, >=99.8%, contains 40-150 ppm amylene as stabilizer; Dichloromethane, anhydrous, contains 40-150 ppm amylene as stabilizer, ZerO2(TM), >=99.8%; Dichloromethane, biotech. grade, 99.9%, contains 40-150 ppm amylene as stabilizer; Dichloromethane, contains 40-150 ppm amylene as stabilizer, ACS reagent, >=99.5%; Dichloromethane, for HPLC, >=99.9%, contains 40-150 ppm amylene as stabilizer; Dichloromethane, puriss., meets analytical specification of Ph.??Eur., NF, >=99% (GC); Dichloromethane, suitable for 1000 per JIS, >=99.5%, for residue analysis; Methylene Chloride, Pharmaceutical Secondary Standard; Certified Reference Material; Dichloromethane, >=99.9%, capillary GC grade, suitable for environmental analysis, contains amylene as stabilizer; Dichloromethane, ACS spectrophotometric grade, >=99.5%, contains 50-150 ppm amylene as stabilizer; Dichloromethane, HPLC Plus, for HPLC, GC, and residue analysis, >=99.9%, contains 50-150 ppm amylene as stabilizer; Dichloromethane, Laboratory Reagent, >=99.9% (without stabilizer, GC), contains 0.1-0.4% ethanol as stabilizer; Dichloromethane, p.a., ACS reagent, reag. ISO, reag. Ph. Eur., 99.8%, contains 40-60 ppm Amylene; Dichloromethane, puriss. p.a., ACS reagent, reag. ISO, dried, >=99.8% (GC), <=0.001% water; M.C; Residual Solvent Class 2 - Methylene chloride, United States Pharmacopeia (USP) Reference Standard
Search Links to Other Online Resources (may not be available):
- Google Structure Search
- EPA CompTox Database
- Chemical Entities of Biological Interest (ChEBI)
- NIH ChemIDplus - A TOXNET DATABASE
- The Human Metabolome Database (HMDB)
- OECD eChemPortal
- Google Scholar
+==================================================+
|| Molecular Calculation ||
+==================================================+
Id = 46454
NWOutput = Link to NWChem Output (download)
Datafiles:
lumo-restricted.cube-746929-2018-4-25-22:38:5 (download)
homo-restricted.cube-746929-2018-4-25-22:38:5 (download)
mo_orbital_nwchemarrows-we23441.out-631319-2019-9-22-5:37:19 (download)
image_resset: api/image_reset/46454
Calculation performed by Eric Bylaska - we29676.emsl.pnl.gov
Numbers of cpus used for calculation = 6
Calculation walltime = 393.000000 seconds (0 days 0 hours 6 minutes 33 seconds)
+----------------+
| Energetic Data |
+----------------+
Id = 46454
iupac = dichloromethane
mformula = C1Cl2H2
inchi = InChI=1S/CH2Cl2/c2-1-3/h1H2
inchikey = YMWUJEATGCHHMB-UHFFFAOYSA-N
esmiles = ClCCl theory{dft} xc{pbe0} basis{6-311++G(2d,2p)} solvation_type{COSMO} ^{0}
calculation_type = ovc
theory = dft
xc = pbe0
basis = 6-311++G(2d,2p)
charge,mult = 0 1
energy = -959.393999 Hartrees
enthalpy correct.= 0.034081 Hartrees
entropy = 65.778 cal/mol-K
solvation energy = -1.599 kcal/mol solvation_type = COSMO
Sitkoff cavity dispersion = 1.861 kcal/mol
Honig cavity dispersion = 5.003 kcal/mol
ASA solvent accesible surface area = 200.122 Angstrom2
ASA solvent accesible volume = 200.900 Angstrom3
+-----------------+
| Structural Data |
+-----------------+
JSmol: an open-source HTML5 viewer for chemical structures in 3D
Number of Atoms = 5
Units are Angstrom for bonds and degrees for angles
Type I J K L M Value
----------- ----- ----- ----- ----- ----- ----------
1 Stretch C1 Cl2 1.77047
2 Stretch C1 Cl3 1.77047
3 Stretch C1 H4 1.08346
4 Stretch C1 H5 1.08346
5 Bend Cl2 C1 Cl3 112.86776
6 Bend Cl2 C1 H4 108.06614
7 Bend Cl2 C1 H5 108.06623
8 Bend Cl3 C1 H4 108.06621
9 Bend Cl3 C1 H5 108.06619
10 Bend H4 C1 H5 111.76526
+-----------------+
| Eigenvalue Data |
+-----------------+
Id = 46454
iupac = dichloromethane
mformula = C1Cl2H2
InChI = InChI=1S/CH2Cl2/c2-1-3/h1H2
smiles = ClCCl
esmiles = ClCCl theory{dft} xc{pbe0} basis{6-311++G(2d,2p)} solvation_type{COSMO} ^{0}
theory = dft
xc = pbe0
basis = 6-311++G(2d,2p)
charge = 0
mult = 1
solvation_type = COSMO
twirl webpage = TwirlMol Link
image webpage = GIF Image Link
Eigevalue Spectra
---- ---- 67.31 eV
-- -- -- -
--- -- ---
---- ----
---- ----
----------
---- ----
---- ----
----------
---- ----
----------
---- ----
----------
----------
---- ----
---- ----
---- ----
- - - - --
7 - - - -
-- -- -- -
----------
----------
-- -- -- -
7 - - - -
- - - - --
---------- LUMO= -0.45 eV
HOMO= -8.88 eV ++++ ++++
++++ ++++
++++ ++++
++++++++++
++++++++++
++++++++++
-25.55 eV ++++++++++
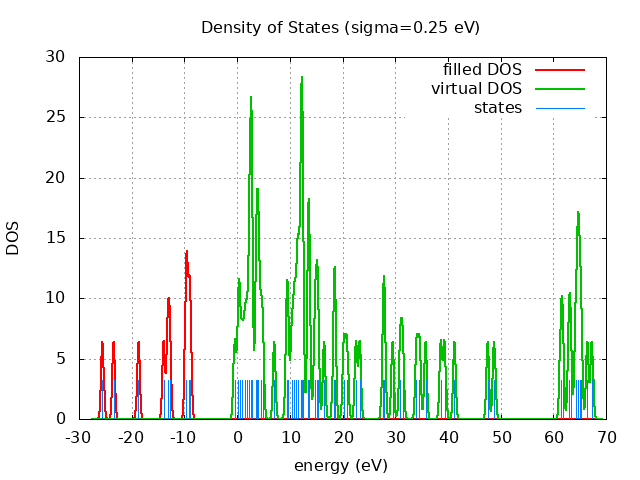
spin eig occ ---------------------------- restricted -25.55 2.00 restricted -23.44 2.00 restricted -18.73 2.00 restricted -13.93 2.00 restricted -13.15 2.00 restricted -12.80 2.00 restricted -9.65 2.00 restricted -9.62 2.00 restricted -9.14 2.00 restricted -8.88 2.00 restricted -0.45 0.00 restricted 0.22 0.00 restricted 0.47 0.00 restricted 0.97 0.00 restricted 1.46 0.00 restricted 1.86 0.00 restricted 2.27 0.00 restricted 2.58 0.00 restricted 2.62 0.00 restricted 2.68 0.00 restricted 2.76 0.00 restricted 3.59 0.00 restricted 3.70 0.00 restricted 4.01 0.00 restricted 4.05 0.00 restricted 4.52 0.00 restricted 4.89 0.00 restricted 7.04 0.00 restricted 9.40 0.00 restricted 9.62 0.00 restricted 10.35 0.00 restricted 10.78 0.00 restricted 11.10 0.00 restricted 11.54 0.00 restricted 11.58 0.00 restricted 12.04 0.00 restricted 12.24 0.00 restricted 12.26 0.00 restricted 12.30 0.00 restricted 12.44 0.00 restricted 13.52 0.00 restricted 13.56 0.00 restricted 13.70 0.00 restricted 14.77 0.00 restricted 15.11 0.00 restricted 15.33 0.00 restricted 16.51 0.00 restricted 18.44 0.00 restricted 18.51 0.00 restricted 20.27 0.00 restricted 20.84 0.00 restricted 22.55 0.00 restricted 23.28 0.00 restricted 27.75 0.00 restricted 27.94 0.00 restricted 29.52 0.00 restricted 30.94 0.00 restricted 31.40 0.00 restricted 34.04 0.00 restricted 34.60 0.00 restricted 35.78 0.00 restricted 38.66 0.00 restricted 39.36 0.00 restricted 41.24 0.00 restricted 47.55 0.00 restricted 48.77 0.00 restricted 61.43 0.00 restricted 61.76 0.00 restricted 62.96 0.00 restricted 63.28 0.00 restricted 64.23 0.00 restricted 64.65 0.00 restricted 64.66 0.00 restricted 65.00 0.00 restricted 65.30 0.00 restricted 66.46 0.00 restricted 67.31 0.00
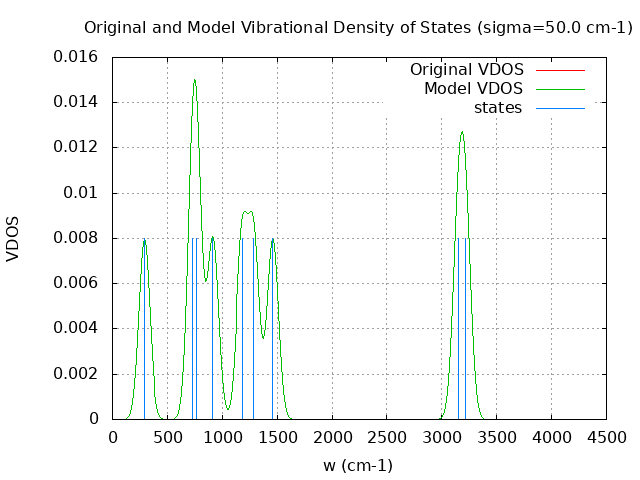
+----------------------------------------+ | Vibrational Density of States Analysis | +----------------------------------------+ Total number of frequencies = 15 Total number of negative frequencies = 0 Number of lowest frequencies = 1 (frequency threshold = 500 ) Exact dos norm = 9.000 Generating vibrational DOS Generating model vibrational DOS to have a proper norm 10.00 9.00 1.00 9.00 50.00 9.00 1.00 9.00 100.00 9.00 1.00 9.00 Generating IR Spectra Writing vibrational density of states (DOS) to vdos.dat Writing model vibrational density of states (DOS_FIXED) to vdos-model.dat Writing IR spectra to irdos.dat Temperature= 298.15 zero-point correction to energy = 18.572 kcal/mol ( 0.029597) vibrational contribution to enthalpy correction = 19.018 kcal/mol ( 0.030306) vibrational contribution to Entropy = 2.204 cal/mol-k hindered rotor enthalpy correction = 0.000 kcal/mol ( 0.000000) hindered rotor entropy correction = 0.000 cal/mol-k
DOS sigma = 10.000000
- vibrational DOS enthalpy correction = 0.030307 kcal/mol ( 19.018 kcal/mol)
- model vibrational DOS enthalpy correction = 0.030305 kcal/mol ( 19.017 kcal/mol)
- vibrational DOS Entropy = 0.000004 ( 2.206 cal/mol-k)
- model vibrational DOS Entropy = 0.000004 ( 2.204 cal/mol-k)
- original gas Energy = -959.393999 (-602028.819 kcal/mol)
- original gas Enthalpy = -959.359918 (-602007.433 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -959.359918 (-602007.433 kcal/mol, delta= 0.000)
- model DOS gas Enthalpy = -959.359920 (-602007.434 kcal/mol, delta= -0.001)
- original gas Entropy = 0.000105 ( 65.778 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000105 ( 65.780 cal/mol-k,delta= 0.002)
- model DOS gas Entropy = 0.000105 ( 65.778 cal/mol-k,delta= -0.000)
- original gas Free Energy = -959.391172 (-602027.045 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -959.391172 (-602027.045 kcal/mol, delta= -0.000)
- model DOS gas Free Energy = -959.391173 (-602027.045 kcal/mol, delta= -0.001)
- original sol Free Energy = -959.393720 (-602028.644 kcal/mol)
- unadjusted DOS sol Free Energy = -959.393721 (-602028.644 kcal/mol)
- model DOS sol Free Energy = -959.393721 (-602028.645 kcal/mol)
DOS sigma = 50.000000
- vibrational DOS enthalpy correction = 0.030315 kcal/mol ( 19.023 kcal/mol)
- model vibrational DOS enthalpy correction = 0.030315 kcal/mol ( 19.023 kcal/mol)
- vibrational DOS Entropy = 0.000004 ( 2.252 cal/mol-k)
- model vibrational DOS Entropy = 0.000004 ( 2.252 cal/mol-k)
- original gas Energy = -959.393999 (-602028.819 kcal/mol)
- original gas Enthalpy = -959.359918 (-602007.433 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -959.359910 (-602007.428 kcal/mol, delta= 0.005)
- model DOS gas Enthalpy = -959.359910 (-602007.428 kcal/mol, delta= 0.005)
- original gas Entropy = 0.000105 ( 65.778 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000105 ( 65.827 cal/mol-k,delta= 0.049)
- model DOS gas Entropy = 0.000105 ( 65.827 cal/mol-k,delta= 0.049)
- original gas Free Energy = -959.391172 (-602027.045 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -959.391186 (-602027.054 kcal/mol, delta= -0.009)
- model DOS gas Free Energy = -959.391186 (-602027.054 kcal/mol, delta= -0.009)
- original sol Free Energy = -959.393720 (-602028.644 kcal/mol)
- unadjusted DOS sol Free Energy = -959.393735 (-602028.653 kcal/mol)
- model DOS sol Free Energy = -959.393735 (-602028.653 kcal/mol)
DOS sigma = 100.000000
- vibrational DOS enthalpy correction = 0.030338 kcal/mol ( 19.037 kcal/mol)
- model vibrational DOS enthalpy correction = 0.030340 kcal/mol ( 19.039 kcal/mol)
- vibrational DOS Entropy = 0.000004 ( 2.422 cal/mol-k)
- model vibrational DOS Entropy = 0.000004 ( 2.426 cal/mol-k)
- original gas Energy = -959.393999 (-602028.819 kcal/mol)
- original gas Enthalpy = -959.359918 (-602007.433 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -959.359887 (-602007.413 kcal/mol, delta= 0.020)
- model DOS gas Enthalpy = -959.359885 (-602007.412 kcal/mol, delta= 0.021)
- original gas Entropy = 0.000105 ( 65.778 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000105 ( 65.996 cal/mol-k,delta= 0.218)
- model DOS gas Entropy = 0.000105 ( 66.000 cal/mol-k,delta= 0.222)
- original gas Free Energy = -959.391172 (-602027.045 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -959.391244 (-602027.090 kcal/mol, delta= -0.045)
- model DOS gas Free Energy = -959.391243 (-602027.090 kcal/mol, delta= -0.045)
- original sol Free Energy = -959.393720 (-602028.644 kcal/mol)
- unadjusted DOS sol Free Energy = -959.393793 (-602028.689 kcal/mol)
- model DOS sol Free Energy = -959.393792 (-602028.689 kcal/mol)
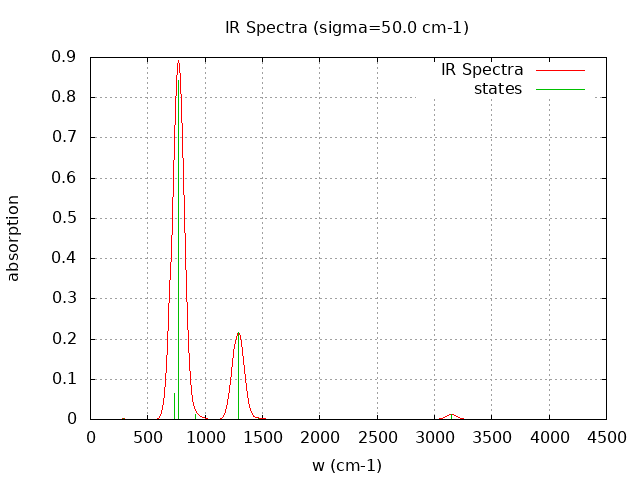
Normal Mode Frequency (cm-1) IR Intensity (arbitrary)
----------- ---------------- ------------------------
1 -0.000 4.359
2 -0.000 0.356
3 -0.000 0.222
4 -0.000 0.089
5 0.000 0.339
6 0.000 0.297
7 289.020 0.160
8 732.070 7.958
9 767.370 105.509
10 913.530 1.540
11 1182.270 0.006
12 1288.140 27.117
13 1461.430 0.454
14 3148.430 1.582
15 3215.560 0.012
No Hindered Rotor Data
+-------------------------------------+
| Reactions Contained in the Database |
+-------------------------------------+
Reactions Containing INCHIKEY = YMWUJEATGCHHMB-UHFFFAOYSA-N
Reactionid Erxn(gas) Hrxn(gas) Grxn(gas) delta_Solv Grxn(aq) ReactionType Reaction
20969 -37.801 -38.130 -37.223 -1.996 -39.219 AB + CD --> AD + BC "ClCCl + OCl --> ClC(Cl)Cl + O"
20890 -433.228 -427.803 -428.852 176.882 -54.770 AB + C --> AC + B "chloroform + 2 SHE + [H+] --> methylene chloride + chloride"
20542 -1.378 -2.426 -1.292 -20.703 -21.995 AB + CD --> AD + BC "ClCCl + [O-]Cl --> ClC(Cl)Cl + [OH-]"
20266 -10.929 -10.681 -10.495 -0.748 -11.243 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
20265 -10.929 -10.681 -10.495 -0.748 -11.243 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
20264 -10.929 -10.681 -10.495 -0.748 -11.243 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
20263 -10.929 -10.681 -10.495 -0.748 -11.243 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
19844 14.957 15.791 15.143 -1.902 13.241 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
19843 14.957 15.791 15.143 -1.902 13.241 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
19842 14.957 15.791 15.143 -1.902 13.241 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
19841 14.957 15.791 15.143 -1.902 13.241 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
19723 11.401 12.524 11.851 -2.271 9.580 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
19722 11.401 12.524 11.851 -2.271 9.580 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
19721 11.401 12.524 11.851 -2.271 9.580 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
19720 11.401 12.524 11.851 -2.271 9.580 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
19455 -10.930 -10.693 -10.537 -0.679 -11.217 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
19454 -10.930 -10.693 -10.537 -0.679 -11.217 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
19453 -10.930 -10.693 -10.537 -0.679 -11.217 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
19452 -10.930 -10.693 -10.537 -0.679 -11.217 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
19396 -241.520 -234.305 -231.635 31.503 -2.932 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> CCl + [Cl] ^{-2} mult{2}"
19395 -268.394 -264.957 -269.066 158.435 86.569 AB + C --> AC + B "ClC(Cl)Cl ^{1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> ClCCl ^{-1} mult{2} + [Cl] ^{1}"
19365 -398.867 -393.456 -388.708 252.887 -37.220 A + B --> AB "Cl[CH]Cl ^{-1} + [H+] ^{1} + [SHE] --> ClCCl ^{-1} mult{2}"
19333 -8.615 -9.624 -14.807 -77.587 6.207 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
19332 -8.615 -9.624 -14.807 -77.587 6.207 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
19331 7.259 6.837 1.624 -16.540 -14.916 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
19330 7.259 6.837 1.624 -16.540 -14.916 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
19329 129.646 134.661 140.040 -132.810 7.230 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [OH-] ^{-1} --> OCCl + [Cl] ^{-2} mult{2}"
19328 167.244 170.768 175.471 -154.768 20.702 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [SH-] ^{-1} --> SCCl + [Cl] ^{-2} mult{2}"
19302 10.913 11.887 11.209 -2.273 8.936 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
19301 10.913 11.887 11.209 -2.273 8.936 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
19300 10.913 11.887 11.209 -2.273 8.936 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
19299 10.913 11.887 11.209 -2.273 8.936 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
18676 14.957 15.802 15.185 -1.971 13.214 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
18675 14.957 15.802 15.185 -1.971 13.214 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
18674 14.957 15.802 15.185 -1.971 13.214 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
18673 14.957 15.802 15.185 -1.971 13.214 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
18553 -25.182 -23.883 -22.102 -8.504 -30.605 AB + C --> AC + B "ClCCl theory{mp2} + [SH-] theory{mp2} --> SCCl theory{mp2} + [Cl-] theory{mp2}"
18168 -576603.303 -576614.108 -576627.188 -153.568 -576780.756 AB --> A + B "Methylene chloride theory{dft} xc{m06-2x} --> [CH2+]Cl theory{dft} xc{m06-2x} + [Cl-] theory{dft} xc{m06-2x}"
18167 -576603.303 -576614.108 -576627.188 -153.568 -576780.756 AB --> A + B "Methylene chloride theory{dft} xc{m06-2x} --> [CH2+]Cl theory{dft} xc{m06-2x} + [Cl-] theory{dft} xc{m06-2x}"
17578 196.479 194.996 186.888 -150.243 36.646 AB --> A + B "Methylene chloride theory{dft} xc{pbe} --> [CH2+]Cl theory{dft} xc{pbe} + [Cl-] theory{dft} xc{pbe}"
17577 196.479 194.996 186.888 -150.243 36.646 AB --> A + B "Methylene chloride theory{dft} xc{pbe} --> [CH2+]Cl theory{dft} xc{pbe} + [Cl-] theory{dft} xc{pbe}"
17576 -10.929 -10.693 -10.537 -0.719 -11.257 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
17575 -10.929 -10.693 -10.537 -0.719 -11.257 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
17574 -10.929 -10.693 -10.537 -0.719 -11.257 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
17573 -10.929 -10.693 -10.537 -0.719 -11.257 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
17567 -380.574 -373.117 -364.961 310.285 -54.676 A + B --> AB "Cl[CH]Cl ^{-1} xc{pbe} + [H+] xc{pbe} --> C(Cl)Cl xc{pbe}"
17539 2.248 3.142 2.595 -1.502 1.093 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
17538 2.248 3.142 2.595 -1.502 1.093 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
17537 2.248 3.142 2.595 -1.502 1.093 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
17536 2.248 3.142 2.595 -1.502 1.093 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
17530 -21.993 -20.832 -19.957 -8.455 -28.412 AB + C --> AC + B "ClCCl xc{pbe} + [SH-] xc{pbe} --> SCCl xc{pbe} + [Cl-] xc{pbe}"
17498 -83.637 -78.380 -68.067 2.824 -65.243 AB + C --> ACB "[CH]Cl xc{pbe} + Cl xc{pbe} --> C(Cl)Cl xc{pbe}"
17497 -83.637 -78.380 -68.067 2.824 -65.243 AB + C --> ACB "[CH]Cl xc{pbe} + Cl xc{pbe} --> C(Cl)Cl xc{pbe}"
17489 -409.713 -402.931 -395.260 256.230 -40.430 A + B --> AB "Cl[CH]Cl xc{pbe} + SHE xc{pbe} + [H+] xc{pbe} --> C(Cl)Cl xc{pbe}"
17279 165.679 169.211 173.843 -154.659 19.183 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [SH-] ^{-1} --> SCCl + [Cl] ^{-2} mult{2}"
17276 170.107 169.191 160.499 -29.522 130.977 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{1}"
17275 170.107 169.191 160.499 -29.522 130.977 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{1}"
17274 -237.308 -231.418 -232.490 182.498 147.208 AB + C --> AC + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> CCl ^{-1} mult{2} + [Cl] ^{1}"
17272 -397.302 -391.899 -387.080 252.778 -35.701 A + B --> AB "Cl[CH]Cl ^{-1} + [H+] ^{1} + [SHE] --> ClCCl ^{-1} mult{2}"
17269 154.233 152.730 144.068 -90.568 152.100 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{1}"
17268 154.233 152.730 144.068 -90.568 152.100 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{1}"
17253 -243.085 -235.862 -233.263 31.612 -4.451 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> CCl + [Cl] ^{-2} mult{2}"
17248 -266.828 -263.400 -267.438 158.326 88.088 AB + C --> AC + B "ClC(Cl)Cl ^{1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> ClCCl ^{-1} mult{2} + [Cl] ^{1}"
17247 -10.180 -11.181 -16.435 -77.477 4.688 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
17246 -10.180 -11.181 -16.435 -77.477 4.688 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
17245 128.080 133.104 138.412 -132.701 5.711 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [OH-] ^{-1} --> OCCl + [Cl] ^{-2} mult{2}"
17237 5.694 5.280 -0.004 -16.431 -16.435 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
17236 5.694 5.280 -0.004 -16.431 -16.435 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
16987 12.320 13.475 12.873 0.000 12.873 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
16986 12.320 13.475 12.873 0.000 12.873 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
16985 12.320 13.475 12.873 0.000 12.873 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
16984 12.320 13.475 12.873 0.000 12.873 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
16848 -8.161 -7.944 -7.843 0.000 -7.843 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
16847 -8.161 -7.944 -7.843 0.000 -7.843 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
16846 -8.161 -7.944 -7.843 0.000 -7.843 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
16845 -8.161 -7.944 -7.843 0.000 -7.843 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
16827 -0.359 0.928 0.336 0.000 0.336 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
16826 -0.359 0.928 0.336 0.000 0.336 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
16825 -0.359 0.928 0.336 0.000 0.336 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
16824 -0.359 0.928 0.336 0.000 0.336 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
16812 -384.051 -376.707 -368.607 0.000 -368.607 A + B --> AB "Cl[CH]Cl ^{-1} theory{pspw4} + [H+] theory{pspw4} --> C(Cl)Cl theory{pspw4}"
16811 -410.274 -403.409 -395.745 0.000 -297.145 A + B --> AB "Cl[CH]Cl theory{pspw4} + SHE theory{pspw4} + [H+] theory{pspw4} --> C(Cl)Cl theory{pspw4}"
16782 8.603 9.585 8.909 -2.253 6.656 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
16781 8.603 9.585 8.909 -2.253 6.656 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
16780 8.603 9.585 8.909 -2.253 6.656 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
16779 8.603 9.585 8.909 -2.253 6.656 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
15642 -385.993 -378.292 -370.069 315.103 -54.966 A + B --> AB "Cl[CH]Cl ^{-1} theory{ccsd(t)} + [H+] theory{ccsd(t)} --> C(Cl)Cl theory{ccsd(t)}"
15574 -415.194 -408.127 -400.463 256.150 -45.714 A + B --> AB "Cl[CH]Cl mult{2} theory{ccsd(t)} + [H+] theory{ccsd(t)} + SHE theory{ccsd(t)} --> C(Cl)Cl theory{ccsd(t)}"
15549 -435.796 -430.369 -431.416 176.922 -57.294 AB + C --> AC + B "chloroform + 2 SHE + [H+] --> methylene chloride + chloride"
15542 -35.233 -35.564 -34.659 -2.036 -36.695 AB + CD --> AD + BC "ClCCl + OCl --> ClC(Cl)Cl + O"
15465 1.190 0.140 1.272 -20.743 -19.471 AB + CD --> AD + BC "ClCCl + [O-]Cl --> ClC(Cl)Cl + [OH-]"
15413 409.886 406.586 397.929 -158.712 239.217 AB --> A + B "ClCCl --> [CH2]Cl ^{-1} + [Cl] ^{1}"
15412 409.886 406.586 397.929 -158.712 239.217 AB --> A + B "ClCCl --> [CH2]Cl ^{-1} + [Cl] ^{1}"
15102 -389.171 -381.592 -373.548 0.000 -373.548 A + B --> AB "Cl[CH]Cl ^{-1} theory{pspw4} xc{pbe0} + [H+] theory{pspw4} xc{pbe0} --> C(Cl)Cl theory{pspw4} xc{pbe0}"
15092 -159.991 -154.679 -146.982 0.000 -146.982 A + B --> AB "Cl[CH]Cl theory{pspw4} xc{pbe0} + [H+] theory{pspw4} xc{pbe0} --> ClCCl ^{+1} theory{pspw4} xc{pbe0}"
15072 -86.420 -80.803 -70.420 2.594 -67.826 AB + C --> ACB "[CH]Cl xc{m06-2x} + Cl xc{m06-2x} --> C(Cl)Cl xc{m06-2x}"
15071 -86.420 -80.803 -70.420 2.594 -67.826 AB + C --> ACB "[CH]Cl xc{m06-2x} + Cl xc{m06-2x} --> C(Cl)Cl xc{m06-2x}"
15070 -87.910 -82.418 -72.049 2.875 -69.174 AB + C --> ACB "[CH]Cl xc{pbe0} + Cl xc{pbe0} --> C(Cl)Cl xc{pbe0}"
15069 -87.910 -82.418 -72.049 2.875 -69.174 AB + C --> ACB "[CH]Cl xc{pbe0} + Cl xc{pbe0} --> C(Cl)Cl xc{pbe0}"
15068 -85.795 -80.530 -70.217 2.804 -67.413 AB + C --> ACB "[CH]Cl xc{pbe} + Cl xc{pbe} --> C(Cl)Cl xc{pbe}"
15067 -85.795 -80.530 -70.217 2.804 -67.413 AB + C --> ACB "[CH]Cl xc{pbe} + Cl xc{pbe} --> C(Cl)Cl xc{pbe}"
15066 -81.215 -75.736 -65.406 2.705 -62.701 AB + C --> ACB "[CH]Cl xc{b3lyp} + Cl xc{b3lyp} --> C(Cl)Cl xc{b3lyp}"
15065 -81.215 -75.736 -65.406 2.705 -62.701 AB + C --> ACB "[CH]Cl xc{b3lyp} + Cl xc{b3lyp} --> C(Cl)Cl xc{b3lyp}"
14887 -384.050 -376.698 -368.596 0.000 -368.596 A + B --> AB "Cl[CH]Cl ^{-1} theory{pspw4} + [H+] theory{pspw4} --> C(Cl)Cl theory{pspw4}"
14886 -383.608 -375.797 -367.702 311.258 -56.444 A + B --> AB "Cl[CH]Cl ^{-1} xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)Cl xc{m06-2x}"
14885 -387.117 -379.431 -371.332 311.359 -59.973 A + B --> AB "Cl[CH]Cl ^{-1} xc{pbe0} + [H+] xc{pbe0} --> C(Cl)Cl xc{pbe0}"
14884 -382.732 -375.266 -367.111 310.265 -56.846 A + B --> AB "Cl[CH]Cl ^{-1} xc{pbe} + [H+] xc{pbe} --> C(Cl)Cl xc{pbe}"
14825 -166.693 -161.385 -153.693 0.000 -153.693 A + B --> AB "Cl[CH]Cl theory{pspw4} + [H+] theory{pspw4} --> ClCCl ^{+1} theory{pspw4}"
14824 -153.096 -148.158 -140.518 187.895 47.377 A + B --> AB "Cl[CH]Cl xc{m06-2x} + [H+] xc{m06-2x} --> ClCCl ^{+1} xc{m06-2x}"
14823 -158.355 -153.185 -145.506 188.066 42.560 A + B --> AB "Cl[CH]Cl xc{pbe0} + [H+] xc{pbe0} --> ClCCl ^{+1} xc{pbe0}"
14822 -165.509 -160.536 -152.919 188.138 35.218 A + B --> AB "Cl[CH]Cl xc{pbe} + [H+] xc{pbe} --> ClCCl ^{+1} xc{pbe}"
14821 -160.642 -155.372 -147.704 187.995 40.291 A + B --> AB "Cl[CH]Cl xc{b3lyp} + [H+] xc{b3lyp} --> ClCCl ^{+1} xc{b3lyp}"
14804 -412.772 -405.610 -397.924 0.000 -299.324 A + B --> AB "Cl[CH]Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} + [H+] theory{pspw4} xc{pbe0} --> C(Cl)Cl theory{pspw4} xc{pbe0}"
14795 -381.927 -374.226 -366.003 315.093 -50.910 A + B --> AB "Cl[CH]Cl ^{-1} xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)Cl xc{b3lyp}"
14790 -410.273 -403.400 -395.734 0.000 -297.134 A + B --> AB "Cl[CH]Cl theory{pspw4} + SHE theory{pspw4} + [H+] theory{pspw4} --> C(Cl)Cl theory{pspw4}"
14789 -416.297 -409.061 -401.390 256.149 -46.640 A + B --> AB "Cl[CH]Cl xc{m06-2x} + SHE xc{m06-2x} + [H+] xc{m06-2x} --> C(Cl)Cl xc{m06-2x}"
14788 -413.728 -406.655 -398.983 256.170 -44.213 A + B --> AB "Cl[CH]Cl xc{pbe0} + SHE xc{pbe0} + [H+] xc{pbe0} --> C(Cl)Cl xc{pbe0}"
14787 -411.870 -405.080 -397.411 256.210 -42.600 A + B --> AB "Cl[CH]Cl xc{pbe} + SHE xc{pbe} + [H+] xc{pbe} --> C(Cl)Cl xc{pbe}"
14786 -416.295 -409.229 -401.565 256.140 -46.826 A + B --> AB "Cl[CH]Cl xc{b3lyp} + SHE xc{b3lyp} + [H+] xc{b3lyp} --> C(Cl)Cl xc{b3lyp}"
14772 -56.853 -54.129 -52.224 13.494 -38.730 AB + C --> AC + B "ClCCl theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> OCCl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
12551 -8.771 -8.553 -8.415 -0.679 -9.094 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
12550 -8.771 -8.553 -8.415 -0.679 -9.094 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
12549 -8.771 -8.553 -8.415 -0.679 -9.094 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
12548 -8.771 -8.553 -8.415 -0.679 -9.094 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
11514 -8.192 -8.044 -8.050 0.000 -8.050 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
11513 -8.192 -8.044 -8.050 0.000 -8.050 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
11512 -8.192 -8.044 -8.050 0.000 -8.050 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
11511 -8.192 -8.044 -8.050 0.000 -8.050 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
7608 199.676 197.902 189.715 -151.012 38.703 AB --> A + B "Methylene chloride theory{dft} xc{m06-2x} --> [CH2+]Cl theory{dft} xc{m06-2x} + [Cl-] theory{dft} xc{m06-2x}"
7607 199.676 197.902 189.715 -151.012 38.703 AB --> A + B "Methylene chloride theory{dft} xc{m06-2x} --> [CH2+]Cl theory{dft} xc{m06-2x} + [Cl-] theory{dft} xc{m06-2x}"
7606 190.538 189.111 181.061 -149.667 31.394 AB --> A + B "Methylene chloride theory{dft} xc{blyp} --> [CH2+]Cl theory{dft} xc{blyp} + [Cl-] theory{dft} xc{blyp}"
7605 190.538 189.111 181.061 -149.667 31.394 AB --> A + B "Methylene chloride theory{dft} xc{blyp} --> [CH2+]Cl theory{dft} xc{blyp} + [Cl-] theory{dft} xc{blyp}"
7604 200.539 198.900 190.723 -151.002 39.721 AB --> A + B "Methylene chloride theory{dft} xc{pbe0} --> [CH2+]Cl theory{dft} xc{pbe0} + [Cl-] theory{dft} xc{pbe0}"
7603 200.539 198.900 190.723 -151.002 39.721 AB --> A + B "Methylene chloride theory{dft} xc{pbe0} --> [CH2+]Cl theory{dft} xc{pbe0} + [Cl-] theory{dft} xc{pbe0}"
7602 198.637 197.145 189.039 -150.223 38.816 AB --> A + B "Methylene chloride theory{dft} xc{pbe} --> [CH2+]Cl theory{dft} xc{pbe} + [Cl-] theory{dft} xc{pbe}"
7601 198.637 197.145 189.039 -150.223 38.816 AB --> A + B "Methylene chloride theory{dft} xc{pbe} --> [CH2+]Cl theory{dft} xc{pbe} + [Cl-] theory{dft} xc{pbe}"
7598 193.433 191.868 183.744 -150.394 33.350 AB --> A + B "Methylene chloride theory{dft} xc{b3lyp} --> [CH2+]Cl theory{dft} xc{b3lyp} + [Cl-] theory{dft} xc{b3lyp}"
7597 193.433 191.868 183.744 -150.394 33.350 AB --> A + B "Methylene chloride theory{dft} xc{b3lyp} --> [CH2+]Cl theory{dft} xc{b3lyp} + [Cl-] theory{dft} xc{b3lyp}"
7532 -175.223 -174.255 -175.809 122.433 -53.376 AB + C --> AC + B "ClCCl + O=C([O-])[O-] --> O=C([O-])OCCl + [Cl-]"
7463 12.800 13.653 13.035 -1.991 11.044 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
7462 12.800 13.653 13.035 -1.991 11.044 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
7461 12.800 13.653 13.035 -1.991 11.044 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
7460 12.800 13.653 13.035 -1.991 11.044 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
7259 0.090 0.993 0.444 -1.522 -1.077 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
7258 0.090 0.993 0.444 -1.522 -1.077 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
7257 0.090 0.993 0.444 -1.522 -1.077 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
7256 0.090 0.993 0.444 -1.522 -1.077 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
5656 -1.377 -2.440 -1.308 -20.773 -22.081 AB + CD --> AD + BC "ClCCl + [O-]Cl --> ClC(Cl)Cl + [OH-]"
5655 -4.410 -4.958 -4.069 -21.638 -25.707 AB + CD --> AD + BC "CCl + [O-]Cl --> ClCCl + [OH-]"
5026 -8.771 -8.554 -8.397 -0.679 -9.076 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
5025 -8.771 -8.554 -8.397 -0.679 -9.076 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
5024 -8.771 -8.554 -8.397 -0.679 -9.076 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
5023 -3.569 -3.781 -4.421 0.000 -4.421 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} xc{pbe0} + methanol theory{pspw} xc{pbe0} --> C(Cl)O theory{pspw} xc{pbe0} + CCl theory{pspw} xc{pbe0}"
5022 -3.569 -3.781 -4.421 0.000 -4.421 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} xc{pbe0} + methanol theory{pspw} xc{pbe0} --> C(Cl)O theory{pspw} xc{pbe0} + CCl theory{pspw} xc{pbe0}"
5021 -3.569 -3.781 -4.421 0.000 -4.421 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} xc{pbe0} + methanol theory{pspw} xc{pbe0} --> C(Cl)O theory{pspw} xc{pbe0} + CCl theory{pspw} xc{pbe0}"
4819 -8.772 -8.543 -8.387 -0.700 -9.086 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
4818 -8.772 -8.543 -8.387 -0.700 -9.086 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
4817 -8.772 -8.543 -8.387 -0.700 -9.086 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
4805 -3.424 -3.633 -4.407 0.000 -4.407 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} xc{pbe0} + methanol theory{pspw4} xc{pbe0} --> C(Cl)O theory{pspw4} xc{pbe0} + CCl theory{pspw4} xc{pbe0}"
4804 -3.424 -3.633 -4.407 0.000 -4.407 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} xc{pbe0} + methanol theory{pspw4} xc{pbe0} --> C(Cl)O theory{pspw4} xc{pbe0} + CCl theory{pspw4} xc{pbe0}"
4803 -3.424 -3.633 -4.407 0.000 -4.407 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} xc{pbe0} + methanol theory{pspw4} xc{pbe0} --> C(Cl)O theory{pspw4} xc{pbe0} + CCl theory{pspw4} xc{pbe0}"
3245 -8.202 -8.004 -7.946 0.000 -7.946 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
3244 -8.202 -8.004 -7.946 0.000 -7.946 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
3243 -8.202 -8.004 -7.946 0.000 -7.946 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
3242 -8.202 -8.004 -7.946 0.000 -7.946 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
3213 -8.161 -7.953 -7.854 0.000 -7.854 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
3212 -8.161 -7.953 -7.854 0.000 -7.854 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
3211 -8.161 -7.953 -7.854 0.000 -7.854 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
2793 -37.800 -38.144 -37.239 -2.066 -39.305 AB + CD --> AD + BC "ClCCl + OCl --> ClC(Cl)Cl + O"
2695 13.204 7.025 9.651 3.076 12.727 ABC + DE --> DBE + AC "ClCCl + ClCl --> ClC(Cl)(Cl)Cl + [HH]"
2559 -0.016 0.989 0.411 -1.472 -1.061 AB + CD --> AD + BC "SC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CS theory{ccsd(t)}"
2558 -0.016 0.989 0.411 -1.472 -1.061 AB + CD --> AD + BC "SC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CS theory{ccsd(t)}"
2557 -0.016 0.989 0.411 -1.472 -1.061 AB + CD --> AD + BC "SC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CS theory{ccsd(t)}"
2556 -1.194 -0.205 -0.871 -1.703 -2.574 AB + CD --> AD + BC "SC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CS xc{m06-2x}"
2555 -1.194 -0.205 -0.871 -1.703 -2.574 AB + CD --> AD + BC "SC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CS xc{m06-2x}"
2554 -1.194 -0.205 -0.871 -1.703 -2.574 AB + CD --> AD + BC "SC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CS xc{m06-2x}"
2553 -1.418 -0.417 -0.991 -1.482 -2.473 AB + CD --> AD + BC "SC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CS xc{b3lyp}"
2552 -1.418 -0.417 -0.991 -1.482 -2.473 AB + CD --> AD + BC "SC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CS xc{b3lyp}"
2551 -1.418 -0.417 -0.991 -1.482 -2.473 AB + CD --> AD + BC "SC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CS xc{b3lyp}"
2550 0.090 0.996 0.447 -1.513 -1.065 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
2549 0.090 0.996 0.447 -1.513 -1.065 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
2548 0.090 0.996 0.447 -1.513 -1.065 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
2547 -0.359 0.937 0.348 0.000 0.348 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
2546 -0.359 0.937 0.348 0.000 0.348 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
2545 -0.359 0.937 0.348 0.000 0.348 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
2529 11.332 12.316 11.657 -2.234 9.422 AB + CD --> AD + BC "OC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CO theory{ccsd(t)}"
2528 11.332 12.316 11.657 -2.234 9.422 AB + CD --> AD + BC "OC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CO theory{ccsd(t)}"
2527 11.332 12.316 11.657 -2.234 9.422 AB + CD --> AD + BC "OC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CO theory{ccsd(t)}"
2526 11.402 12.523 11.852 -2.321 9.531 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
2525 11.402 12.523 11.852 -2.321 9.531 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
2524 11.402 12.523 11.852 -2.321 9.531 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
2477 10.913 11.890 11.209 -2.175 9.035 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
2476 10.913 11.890 11.209 -2.175 9.035 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
2475 10.913 11.890 11.209 -2.175 9.035 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
2474 12.800 13.656 13.038 -1.982 11.056 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
2473 12.800 13.656 13.038 -1.982 11.056 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
2472 12.800 13.656 13.038 -1.982 11.056 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
2471 12.321 13.484 12.884 0.000 12.884 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
2470 12.321 13.484 12.884 0.000 12.884 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
2469 12.321 13.484 12.884 0.000 12.884 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
2166 3.602 2.773 3.872 0.117 3.988 AB + CD --> AD + BC "ClCCl + O --> Cl + OCCl"
2165 -21.277 -20.050 -18.632 -8.485 -27.118 AB + C --> AC + B "ClCCl xc{pbe0} + [SH-] xc{pbe0} --> SCCl xc{pbe0} + [Cl-] xc{pbe0}"
2164 -40.833 -40.662 -39.999 -2.931 -42.930 AB + CD --> AD + BC "OCl + CCl --> O + ClCCl"
2095 -9.681 -12.393 -21.081 -78.746 -1.227 AB --> A + B "C(Cl)Cl xc{b3lyp} + [SHE] xc{b3lyp} --> [CH2]Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
1790 -8.202 -7.988 -7.929 0.000 -7.929 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
1789 -8.202 -7.988 -7.929 0.000 -7.929 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
1788 -8.202 -7.988 -7.929 0.000 -7.929 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
1653 -8.202 -7.988 -7.929 0.000 -7.929 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} + methanol theory{pspw} --> C(Cl)O theory{pspw} + CCl theory{pspw}"
1644 -3.424 -3.633 -4.407 0.000 -4.407 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} xc{pbe0} + methanol theory{pspw4} xc{pbe0} --> C(Cl)O theory{pspw4} xc{pbe0} + CCl theory{pspw4} xc{pbe0}"
1642 -3.569 -3.781 -4.421 0.000 -4.421 AB + CD --> AD + BC "C(Cl)Cl theory{pspw} xc{pbe0} + methanol theory{pspw} xc{pbe0} --> C(Cl)O theory{pspw} xc{pbe0} + CCl theory{pspw} xc{pbe0}"
1633 -8.771 -8.554 -8.397 -0.679 -9.076 AB + CD --> AD + BC "C(Cl)Cl xc{pbe0} + methanol xc{pbe0} --> C(Cl)O xc{pbe0} + CCl xc{pbe0}"
1632 -8.772 -8.543 -8.387 -0.700 -9.086 AB + CD --> AD + BC "C(Cl)Cl xc{pbe} + methanol xc{pbe} --> C(Cl)O xc{pbe} + CCl xc{pbe}"
1631 -8.161 -7.953 -7.854 0.000 -7.854 AB + CD --> AD + BC "C(Cl)Cl theory{pspw4} + methanol theory{pspw4} --> C(Cl)O theory{pspw4} + CCl theory{pspw4}"
1574 13.204 7.025 9.651 3.076 12.727 ABC + DE --> DBE + AC "ClCCl + ClCl --> ClC(Cl)(Cl)Cl + [HH]"
1559 -21.432 -20.198 -18.970 -8.454 -27.424 AB + C --> AC + B "ClCCl + [SH-] --> SCCl + [Cl-]"
1549 -40.833 -40.662 -39.999 -2.931 -42.930 AB + CD --> AD + BC "OCl + CCl --> O + ClCCl"
1547 3.602 2.773 3.872 0.117 3.988 AB + CD --> AD + BC "ClCCl + O --> Cl + OCCl"
1518 -0.016 0.989 0.411 -1.472 -1.061 AB + CD --> AD + BC "SC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CS theory{ccsd(t)}"
1515 11.332 12.316 11.657 -2.234 9.422 AB + CD --> AD + BC "OC(Cl)Cl theory{ccsd(t)} + C theory{ccsd(t)} --> ClCCl theory{ccsd(t)} + CO theory{ccsd(t)}"
1490 12.321 13.484 12.884 0.000 12.884 AB + CD --> AD + BC "OC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CO theory{pspw4}"
1485 -1.194 -0.205 -0.871 -1.703 -2.574 AB + CD --> AD + BC "SC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CS xc{m06-2x}"
1484 -1.418 -0.417 -0.991 -1.482 -2.473 AB + CD --> AD + BC "SC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CS xc{b3lyp}"
1483 0.090 0.996 0.447 -1.513 -1.065 AB + CD --> AD + BC "SC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CS theory{dft} xc{pbe}"
1482 -0.359 0.937 0.348 0.000 0.348 AB + CD --> AD + BC "SC(Cl)Cl theory{pspw4} + C theory{pspw4} --> ClCCl theory{pspw4} + CS theory{pspw4}"
1474 11.402 12.523 11.852 -2.321 9.531 AB + CD --> AD + BC "OC(Cl)Cl xc{m06-2x} + C xc{m06-2x} --> ClCCl xc{m06-2x} + CO xc{m06-2x}"
1473 10.913 11.890 11.209 -2.175 9.035 AB + CD --> AD + BC "OC(Cl)Cl xc{b3lyp} + C xc{b3lyp} --> ClCCl xc{b3lyp} + CO xc{b3lyp}"
1472 12.800 13.656 13.038 -1.982 11.056 AB + CD --> AD + BC "OC(Cl)Cl theory{dft} xc{pbe} + C theory{dft} xc{pbe} --> ClCCl theory{dft} xc{pbe} + CO theory{dft} xc{pbe}"
829 -430.196 -425.271 -426.076 177.817 -51.058 AB + C --> AC + B "methylene chloride + 2 SHE + [H+] --> methyl chloride + [Cl-]"
828 -433.230 -427.789 -428.836 176.952 -54.684 AB + C --> AC + B "chloroform + 2 SHE + [H+] --> methylene chloride + chloride"
815 -9.681 -12.393 -21.081 -78.746 -1.227 AB --> A + B "Dichloromethane + SHE --> [CH2]Cl + chloride"
721 3.602 2.772 3.870 0.106 3.977 AB + CD --> AD + BC "ClCCl + O --> Cl + OCCl"
205 -21.834 -20.649 -19.016 -8.475 -27.490 AB + C --> AC + B "ClCCl xc{m06-2x} + [SH-] xc{m06-2x} --> SCCl xc{m06-2x} + [Cl-] xc{m06-2x}"
204 -21.277 -20.047 -18.622 -8.485 -27.107 AB + C --> AC + B "ClCCl xc{pbe0} + [SH-] xc{pbe0} --> SCCl xc{pbe0} + [Cl-] xc{pbe0}"
203 -19.835 -18.683 -17.806 -8.436 -26.242 AB + C --> AC + B "ClCCl xc{pbe} + [SH-] xc{pbe} --> SCCl xc{pbe} + [Cl-] xc{pbe}"
202 -19.789 -18.663 -17.707 -8.477 -26.184 AB + C --> AC + B "ClCCl xc{lda} + [SH-] xc{lda} --> SCCl xc{lda} + [Cl-] xc{lda}"
201 -25.188 -23.892 -22.109 -8.514 -30.623 AB + C --> AC + B "ClCCl theory{mp2} + [SH-] theory{mp2} --> SCCl theory{mp2} + [Cl-] theory{mp2}"
200 -24.571 -23.275 -21.492 -8.514 -30.006 AB + C --> AC + B "ClCCl theory{ccsd(t)} + [SH-] theory{ccsd(t)} --> SCCl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
181 -24.198 -22.902 -21.120 -8.514 -29.633 AB + C --> AC + B "ClCCl + [SH-] --> SCCl + [Cl-]"
170 -56.853 -54.128 -52.223 13.574 -38.649 AB + C --> AC + B "ClCCl theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> OCCl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
169 -59.030 -56.306 -54.401 13.494 -40.907 AB + C --> AC + B "ClCCl + [OH-] --> OCCl + [Cl-]"
162 13.205 7.024 9.655 3.017 12.673 ABC + DE --> DBE + AC "ClCCl + ClCl --> ClC(Cl)(Cl)Cl + [HH]"
11 -37.800 -38.145 -37.239 -2.076 -39.315 AB + CD --> AD + BC "ClCCl + OCl --> ClC(Cl)Cl + O"
10 -40.833 -40.661 -39.998 -2.913 -42.911 AB + CD --> AD + BC "OCl + CCl --> O + ClCCl"
All requests to Arrows were successful.
KEYWORDs -
reaction: :reaction
chinese_room: :chinese_room
molecule: :molecule
nmr: :nmr
predict: :predict
submitesmiles: :submitesmiles
nosubmitmissingesmiles
resubmitmissingesmiles
submitmachines: :submitmachines
useallentries
nomodelcorrect
eigenvalues: :eigenvalues
frequencies: :frequencies
nwoutput: :nwoutput
xyzfile: :xyzfile
alleigs: :alleigs
allfreqs: :allfreqs
reactionenumerate:
energytype:[erxn(gas) hrxn(gas) grxn(gas) delta_solvation grxn(aq)] :energytype
energytype:[kcal/mol kj/mol ev cm-1 ry hartree au] :energytype
tablereactions:
reaction: ... :reaction
reaction: ... :reaction
...
:tablereactions
tablemethods:
method: ... :method
method: ... :method
...
:tablemethods
:reactionenumerate
rotatebonds
xyzinput:
label: :label
xyzdata:
... xyz data ...
:xyzdata
:xyzinput
submitHf: :submitHf
nmrexp: :nmrexp
findreplace: old text | new text :findreplace
listnwjobs
fetchnwjob: :fetchnwjob
pushnwjob: :pushnwjob
printcsv: :printcsv
printeig: :printeig
printfreq: :printfreq
printxyz: :printxyz
printjobinfo: :printjobinfo
printnwout: :printnwout
badids: :badids
hup_string:
database:
table:
request_table:
listallesmiles
queuecheck
This software service and its documentation were developed at the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory, a multiprogram national laboratory, operated for the U.S. Department of Energy by Battelle under Contract Number DE-AC05-76RL01830. Support for this work was provided by the Department of Energy Office of Biological and Environmental Research, and Department of Defense environmental science and technology program (SERDP). THE SOFTWARE SERVICE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE SERVICE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE SERVICE.