 3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual
3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual Results from an EMSL Arrows Calculation
| EMSL Arrows is a revolutionary approach to materials and chemical simulations that uses NWChem and chemical computational databases to make materials and chemical modeling accessible via a broad spectrum of digital communications including posts to web APIs, social networks, and traditional email. |
Molecular modeling software has previously been extremely complex, making it prohibative to all but experts in the field, yet even experts can struggle to perform calculations. This service is designed to be used by experts and non-experts alike. Experts can carry out and keep track of large numbers of complex calculations with diverse levels of theories present in their workflows. Additionally, due to a streamlined and easy-to-use input, non-experts can carry out a wide variety of molecular modeling calculations previously not accessible to them.
The id(s) for emsiles = C1CC2C1CC2 theory{dft} xc{b3lyp} basis{6-311++G(2d,2p)} solvation_type{COSMO} ^{0} are: 43306
Use id=% instead of esmiles to print other entries.
mformula = C6H4
iupac = bicyclo[2.2.0]hexa-1(4),2,5-triene
PubChem = 3030745
PubChem LCSS = 3030745
synonyms = bicyclo[2.2.0]hexa-1(4),2,5-triene; Butalene; AC1MHQKF
Search Links to Other Online Resources (may not be available):
- Google Structure Search
- EPA CompTox Database
- Chemical Entities of Biological Interest (ChEBI)
- NIH ChemIDplus - A TOXNET DATABASE
- The Human Metabolome Database (HMDB)
- OECD eChemPortal
- Google Scholar
+==================================================+
|| Molecular Calculation ||
+==================================================+
Id = 43306
NWOutput = Link to NWChem Output (download)
Datafiles:
homo-restricted.cube-776878-2017-7-7-11:37:1 (download)
lumo-restricted.cube-776878-2017-7-7-11:37:1 (download)
mo_orbital_nwchemarrows-2023-10-13-7-15-179723.out-390214-2023-10-13-7:37:1 (download)
image_resset: api/image_reset/43306
Calculation performed by Eric Bylaska - we16124.emsl.pnl.gov
Numbers of cpus used for calculation = 2
Calculation walltime = 10694.700000 seconds (0 days 2 hours 58 minutes 14 seconds)
+----------------+
| Energetic Data |
+----------------+
Id = 43306
iupac = bicyclo[2.2.0]hexa-1(4),2,5-triene
mformula = C6H4
inchi = InChI=1S/C6H10/c1-2-6-4-3-5(1)6/h5-6H,1-4H2
inchikey = YHCJOCYHUDCVQI-UHFFFAOYSA-N
esmiles = C1CC2C1CC2 theory{dft} xc{b3lyp} basis{6-311++G(2d,2p)} solvation_type{COSMO} ^{0}
calculation_type = ovc
theory = dft
xc = b3lyp
basis = 6-311++G(2d,2p)
charge,mult = 0 1
energy = -230.886723 Hartrees
enthalpy correct.= 0.077982 Hartrees
entropy = 69.808 cal/mol-K
solvation energy = -0.779 kcal/mol solvation_type = COSMO
Sitkoff cavity dispersion = 2.031 kcal/mol
Honig cavity dispersion = 5.854 kcal/mol
ASA solvent accesible surface area = 234.149 Angstrom2
ASA solvent accesible volume = 222.061 Angstrom3
+-----------------+
| Structural Data |
+-----------------+
JSmol: an open-source HTML5 viewer for chemical structures in 3D
Number of Atoms = 10
Units are Angstrom for bonds and degrees for angles
Type I J K L M Value
----------- ----- ----- ----- ----- ----- ----------
1 Stretch C1 C2 1.38960
2 Stretch C1 C6 1.44904
3 Stretch C1 H7 1.08348
4 Stretch C2 C3 1.38987
5 Stretch C2 C5 1.56885
6 Stretch C3 C4 1.44823
7 Stretch C3 H8 1.08333
8 Stretch C4 C5 1.38987
9 Stretch C4 H9 1.08333
10 Stretch C5 C6 1.38961
11 Stretch C6 H10 1.08349
12 Bend C2 C1 C6 92.47167
13 Bend C2 C1 H7 134.14335
14 Bend C6 C1 H7 133.38394
15 Bend C1 C2 C3 175.04188
16 Bend C1 C2 C5 87.52837
17 Bend C3 C2 C5 87.51352
18 Bend C2 C3 C4 92.48631
19 Bend C2 C3 H8 134.13078
20 Bend C4 C3 H8 133.38247
21 Bend C3 C4 C5 92.48754
22 Bend C3 C4 H9 133.38250
23 Bend C5 C4 H9 134.12937
24 Bend C2 C5 C4 87.51253
25 Bend C2 C5 C6 87.53017
26 Bend C4 C5 C6 175.04264
27 Bend C1 C6 C5 92.46962
28 Bend C1 C6 H10 133.38533
29 Bend C5 C6 H10 134.14421
30 Dihedral C1 C2 C3 C4 -0.06050
31 Dihedral C1 C2 C3 H8 179.71549
32 Dihedral C1 C2 C5 C4 -179.92511
33 Dihedral C1 C2 C5 C6 0.09971
34 Dihedral C1 C6 C5 C2 -0.09562
35 Dihedral C1 C6 C5 C4 -0.38260
36 Dihedral C2 C1 C6 C5 0.10796
37 Dihedral C2 C1 C6 H10 -179.58525
38 Dihedral C2 C3 C4 C5 0.08015
39 Dihedral C2 C3 C4 H9 -179.66124
40 Dihedral C2 C5 C4 C3 -0.07100
41 Dihedral C2 C5 C4 H9 179.66714
42 Dihedral C2 C5 C6 H10 179.59367
43 Dihedral C3 C2 C1 C6 -0.10613
44 Dihedral C3 C2 C1 H7 179.54692
45 Dihedral C3 C2 C5 C4 0.07398
46 Dihedral C3 C2 C5 C6 -179.90119
47 Dihedral C3 C4 C5 C6 0.21598
48 Dihedral C4 C3 C2 C5 -0.07100
49 Dihedral C4 C5 C6 H10 179.30669
50 Dihedral C5 C2 C1 C6 -0.09562
51 Dihedral C5 C2 C1 H7 179.55742
52 Dihedral C5 C2 C3 H8 179.70498
53 Dihedral C5 C4 C3 H8 -179.69862
54 Dihedral C5 C6 C1 H7 -179.54947
55 Dihedral C6 C5 C4 H9 179.95412
56 Dihedral H7 C1 C6 H10 0.75732
57 Dihedral H8 C3 C4 H9 0.55999
+-----------------+
| Eigenvalue Data |
+-----------------+
Id = 43306
iupac = bicyclo[2.2.0]hexa-1(4),2,5-triene
mformula = C6H4
InChI = InChI=1S/C6H10/c1-2-6-4-3-5(1)6/h5-6H,1-4H2
smiles = c1cc2c1cc2
esmiles = C1CC2C1CC2 theory{dft} xc{b3lyp} basis{6-311++G(2d,2p)} solvation_type{COSMO} ^{0}
theory = dft
xc = b3lyp
basis = 6-311++G(2d,2p)
charge = 0
mult = 1
solvation_type = COSMO
twirl webpage = TwirlMol Link
image webpage = GIF Image Link
Eigevalue Spectra
---------- 61.67 eV
----------
----------
----------
---- ----
---- ----
----------
- - - - --
--- -- ---
----------
--- -- ---
6 - - - -
-- -- -- -
---- ----
---- ----
--- -- ---
6 - - - -
-- -- -- -
---- ----
--- -- ---
7 - - - -
6 - - - -
7 - - - -
--- -- ---
----------
-- -- -- -
-- -- -- -
7 - - - -
12 - - - -
6 - - - -
-- -- -- - LUMO= -1.43 eV
HOMO= -6.15 eV ++++++++++
++++ ++++
+++ ++ +++
++++ ++++
++++++++++
++++ ++++
++++++++++
++++++++++
-25.47 eV ++++++++++
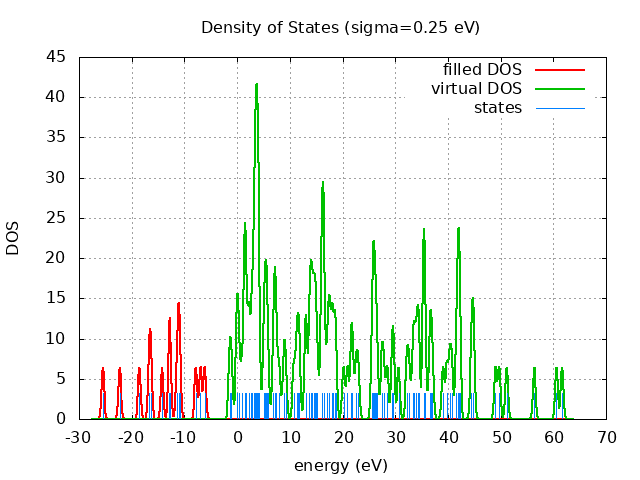
spin eig occ ---------------------------- restricted -25.47 2.00 restricted -22.27 2.00 restricted -18.55 2.00 restricted -16.64 2.00 restricted -16.39 2.00 restricted -14.23 2.00 restricted -12.84 2.00 restricted -12.75 2.00 restricted -11.39 2.00 restricted -11.04 2.00 restricted -10.91 2.00 restricted -7.83 2.00 restricted -6.95 2.00 restricted -6.15 2.00 restricted -1.43 0.00 restricted -1.09 0.00 restricted -0.08 0.00 restricted 0.06 0.00 restricted 0.32 0.00 restricted 0.90 0.00 restricted 1.42 0.00 restricted 1.48 0.00 restricted 1.52 0.00 restricted 1.68 0.00 restricted 2.17 0.00 restricted 2.25 0.00 restricted 2.73 0.00 restricted 2.87 0.00 restricted 3.20 0.00 restricted 3.31 0.00 restricted 3.39 0.00 restricted 3.59 0.00 restricted 3.60 0.00 restricted 3.65 0.00 restricted 3.76 0.00 restricted 3.83 0.00 restricted 4.03 0.00 restricted 4.06 0.00 restricted 4.13 0.00 restricted 5.04 0.00 restricted 5.28 0.00 restricted 5.43 0.00 restricted 5.63 0.00 restricted 5.85 0.00 restricted 6.86 0.00 restricted 7.11 0.00 restricted 7.24 0.00 restricted 7.44 0.00 restricted 7.98 0.00 restricted 8.83 0.00 restricted 9.20 0.00 restricted 10.74 0.00 restricted 11.27 0.00 restricted 11.55 0.00 restricted 11.83 0.00 restricted 13.01 0.00 restricted 13.03 0.00 restricted 13.68 0.00 restricted 13.97 0.00 restricted 13.99 0.00 restricted 14.30 0.00 restricted 14.51 0.00 restricted 14.74 0.00 restricted 14.92 0.00 restricted 15.23 0.00 restricted 16.04 0.00 restricted 16.06 0.00 restricted 16.12 0.00 restricted 16.42 0.00 restricted 16.43 0.00 restricted 16.45 0.00 restricted 17.07 0.00 restricted 17.47 0.00 restricted 17.50 0.00 restricted 18.00 0.00 restricted 18.11 0.00 restricted 18.56 0.00 restricted 18.71 0.00 restricted 20.19 0.00 restricted 20.94 0.00 restricted 21.68 0.00 restricted 21.87 0.00 restricted 22.54 0.00 restricted 22.99 0.00 restricted 25.59 0.00 restricted 25.81 0.00 restricted 25.90 0.00 restricted 26.09 0.00 restricted 26.31 0.00 restricted 26.55 0.00 restricted 27.42 0.00 restricted 27.80 0.00 restricted 28.48 0.00 restricted 29.45 0.00 restricted 29.67 0.00 restricted 30.63 0.00 restricted 32.19 0.00 restricted 32.60 0.00 restricted 33.32 0.00 restricted 33.63 0.00 restricted 33.96 0.00 restricted 34.33 0.00 restricted 34.46 0.00 restricted 35.41 0.00 restricted 35.42 0.00 restricted 35.44 0.00 restricted 35.66 0.00 restricted 36.53 0.00 restricted 36.75 0.00 restricted 37.06 0.00 restricted 39.07 0.00 restricted 39.78 0.00 restricted 40.33 0.00 restricted 40.73 0.00 restricted 41.59 0.00 restricted 41.89 0.00 restricted 42.00 0.00 restricted 42.14 0.00 restricted 42.22 0.00 restricted 44.43 0.00 restricted 44.79 0.00 restricted 44.83 0.00 restricted 49.02 0.00 restricted 49.72 0.00 restricted 51.16 0.00 restricted 56.40 0.00 restricted 60.60 0.00 restricted 61.67 0.00
+----------------------------------------+ | Vibrational Density of States Analysis | +----------------------------------------+ Total number of frequencies = 30 Total number of negative frequencies = 0 Number of lowest frequencies = 2 (frequency threshold = 500 ) Exact dos norm = 24.000 Generating vibrational DOS Generating model vibrational DOS to have a proper norm 10.00 24.00 2.00 24.00 50.00 24.00 2.00 24.00 100.00 23.99 1.99 24.00 Generating IR Spectra Writing vibrational density of states (DOS) to vdos.dat Writing model vibrational density of states (DOS_FIXED) to vdos-model.dat Writing IR spectra to irdos.dat Temperature= 298.15 zero-point correction to energy = 45.397 kcal/mol ( 0.072345) vibrational contribution to enthalpy correction = 46.566 kcal/mol ( 0.074207) vibrational contribution to Entropy = 5.749 cal/mol-k hindered rotor enthalpy correction = 0.000 kcal/mol ( 0.000000) hindered rotor entropy correction = 0.000 cal/mol-k
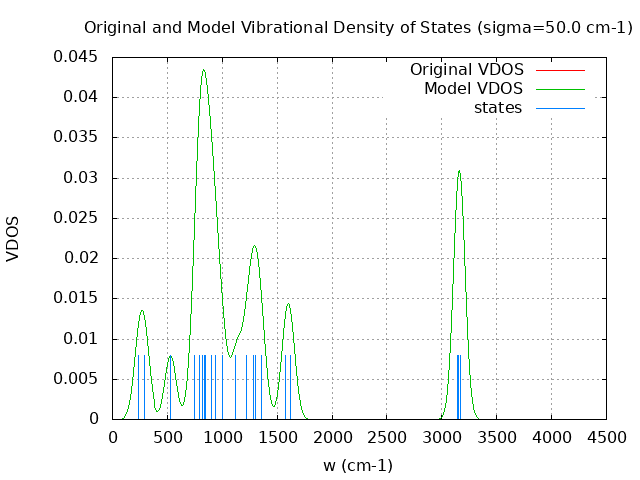
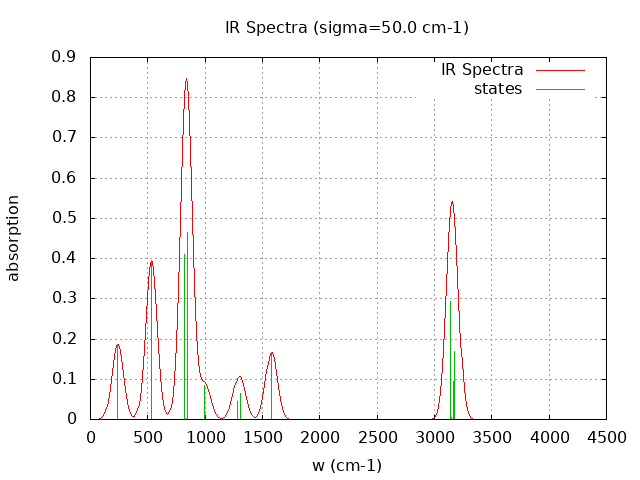
DOS sigma = 10.000000
- vibrational DOS enthalpy correction = 0.074208 kcal/mol ( 46.566 kcal/mol)
- model vibrational DOS enthalpy correction = 0.074205 kcal/mol ( 46.564 kcal/mol)
- vibrational DOS Entropy = 0.000009 ( 5.755 cal/mol-k)
- model vibrational DOS Entropy = 0.000009 ( 5.749 cal/mol-k)
- original gas Energy = -230.886723 (-144883.605 kcal/mol)
- original gas Enthalpy = -230.808741 (-144834.671 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -230.808740 (-144834.670 kcal/mol, delta= 0.001)
- model DOS gas Enthalpy = -230.808744 (-144834.672 kcal/mol, delta= -0.002)
- original gas Entropy = 0.000111 ( 69.808 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000111 ( 69.813 cal/mol-k,delta= 0.005)
- model DOS gas Entropy = 0.000111 ( 69.808 cal/mol-k,delta= -0.000)
- original gas Free Energy = -230.841909 (-144855.484 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -230.841911 (-144855.485 kcal/mol, delta= -0.001)
- model DOS gas Free Energy = -230.841912 (-144855.485 kcal/mol, delta= -0.002)
- original sol Free Energy = -230.843151 (-144856.263 kcal/mol)
- unadjusted DOS sol Free Energy = -230.843152 (-144856.264 kcal/mol)
- model DOS sol Free Energy = -230.843153 (-144856.265 kcal/mol)
DOS sigma = 50.000000
- vibrational DOS enthalpy correction = 0.074229 kcal/mol ( 46.580 kcal/mol)
- model vibrational DOS enthalpy correction = 0.074229 kcal/mol ( 46.580 kcal/mol)
- vibrational DOS Entropy = 0.000009 ( 5.881 cal/mol-k)
- model vibrational DOS Entropy = 0.000009 ( 5.881 cal/mol-k)
- original gas Energy = -230.886723 (-144883.605 kcal/mol)
- original gas Enthalpy = -230.808741 (-144834.671 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -230.808719 (-144834.657 kcal/mol, delta= 0.014)
- model DOS gas Enthalpy = -230.808719 (-144834.657 kcal/mol, delta= 0.014)
- original gas Entropy = 0.000111 ( 69.808 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000111 ( 69.940 cal/mol-k,delta= 0.132)
- model DOS gas Entropy = 0.000111 ( 69.940 cal/mol-k,delta= 0.132)
- original gas Free Energy = -230.841909 (-144855.484 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -230.841950 (-144855.509 kcal/mol, delta= -0.025)
- model DOS gas Free Energy = -230.841950 (-144855.509 kcal/mol, delta= -0.025)
- original sol Free Energy = -230.843151 (-144856.263 kcal/mol)
- unadjusted DOS sol Free Energy = -230.843191 (-144856.288 kcal/mol)
- model DOS sol Free Energy = -230.843191 (-144856.288 kcal/mol)
DOS sigma = 100.000000
- vibrational DOS enthalpy correction = 0.074288 kcal/mol ( 46.616 kcal/mol)
- model vibrational DOS enthalpy correction = 0.074299 kcal/mol ( 46.623 kcal/mol)
- vibrational DOS Entropy = 0.000010 ( 6.326 cal/mol-k)
- model vibrational DOS Entropy = 0.000010 ( 6.345 cal/mol-k)
- original gas Energy = -230.886723 (-144883.605 kcal/mol)
- original gas Enthalpy = -230.808741 (-144834.671 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -230.808660 (-144834.620 kcal/mol, delta= 0.051)
- model DOS gas Enthalpy = -230.808649 (-144834.613 kcal/mol, delta= 0.058)
- original gas Entropy = 0.000111 ( 69.808 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000112 ( 70.385 cal/mol-k,delta= 0.577)
- model DOS gas Entropy = 0.000112 ( 70.404 cal/mol-k,delta= 0.596)
- original gas Free Energy = -230.841909 (-144855.484 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -230.842102 (-144855.605 kcal/mol, delta= -0.121)
- model DOS gas Free Energy = -230.842100 (-144855.604 kcal/mol, delta= -0.120)
- original sol Free Energy = -230.843151 (-144856.263 kcal/mol)
- unadjusted DOS sol Free Energy = -230.843344 (-144856.384 kcal/mol)
- model DOS sol Free Energy = -230.843342 (-144856.383 kcal/mol)
Normal Mode Frequency (cm-1) IR Intensity (arbitrary)
----------- ---------------- ------------------------
1 -0.000 0.266
2 -0.000 0.000
3 -0.000 0.726
4 -0.000 0.000
5 0.000 0.000
6 0.000 0.037
7 237.700 23.521
8 294.540 0.002
9 531.430 49.536
10 747.490 0.006
11 789.370 0.011
12 793.680 0.000
13 822.270 51.497
14 837.440 0.356
15 849.620 58.370
16 899.910 0.002
17 904.220 0.002
18 939.790 0.012
19 998.090 10.505
20 1123.830 0.001
21 1220.630 0.023
22 1282.710 5.671
23 1307.150 8.178
24 1359.460 0.001
25 1578.380 20.931
26 1622.870 0.002
27 3143.540 36.626
28 3147.870 0.614
29 3168.150 11.838
30 3171.020 21.262
No Hindered Rotor Data
+-------------------------------------+
| Reactions Contained in the Database |
+-------------------------------------+
Reactions Containing INCHIKEY = YHCJOCYHUDCVQI-UHFFFAOYSA-N
Reactionid Erxn(gas) Hrxn(gas) Grxn(gas) delta_Solv Grxn(aq) ReactionType Reaction
16994 161.548 152.229 142.265 1.452 143.717 AB + CD --> AD + BC "C1=CC=CC=C1 --> C1=C2C=CC2=C1 + [H][H]"
All requests to Arrows were successful.
KEYWORDs -
reaction: :reaction
chinese_room: :chinese_room
molecule: :molecule
nmr: :nmr
predict: :predict
submitesmiles: :submitesmiles
nosubmitmissingesmiles
resubmitmissingesmiles
submitmachines: :submitmachines
useallentries
nomodelcorrect
eigenvalues: :eigenvalues
frequencies: :frequencies
nwoutput: :nwoutput
xyzfile: :xyzfile
alleigs: :alleigs
allfreqs: :allfreqs
reactionenumerate:
energytype:[erxn(gas) hrxn(gas) grxn(gas) delta_solvation grxn(aq)] :energytype
energytype:[kcal/mol kj/mol ev cm-1 ry hartree au] :energytype
tablereactions:
reaction: ... :reaction
reaction: ... :reaction
...
:tablereactions
tablemethods:
method: ... :method
method: ... :method
...
:tablemethods
:reactionenumerate
rotatebonds
xyzinput:
label: :label
xyzdata:
... xyz data ...
:xyzdata
:xyzinput
submitHf: :submitHf
nmrexp: :nmrexp
findreplace: old text | new text :findreplace
listnwjobs
fetchnwjob: :fetchnwjob
pushnwjob: :pushnwjob
printcsv: :printcsv
printeig: :printeig
printfreq: :printfreq
printxyz: :printxyz
printjobinfo: :printjobinfo
printnwout: :printnwout
badids: :badids
hup_string:
database:
table:
request_table:
listallesmiles
queuecheck
This software service and its documentation were developed at the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory, a multiprogram national laboratory, operated for the U.S. Department of Energy by Battelle under Contract Number DE-AC05-76RL01830. Support for this work was provided by the Department of Energy Office of Biological and Environmental Research, and Department of Defense environmental science and technology program (SERDP). THE SOFTWARE SERVICE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE SERVICE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE SERVICE.