 3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual
3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual Results from an EMSL Arrows Calculation
| EMSL Arrows is a revolutionary approach to materials and chemical simulations that uses NWChem and chemical computational databases to make materials and chemical modeling accessible via a broad spectrum of digital communications including posts to web APIs, social networks, and traditional email. |
Molecular modeling software has previously been extremely complex, making it prohibative to all but experts in the field, yet even experts can struggle to perform calculations. This service is designed to be used by experts and non-experts alike. Experts can carry out and keep track of large numbers of complex calculations with diverse levels of theories present in their workflows. Additionally, due to a streamlined and easy-to-use input, non-experts can carry out a wide variety of molecular modeling calculations previously not accessible to them.
The id(s) for emsiles = [Cl] theory{pspw} xc{b3lyp} basis{100.0 Ry} solvation_type{None} ^{0} mult{2} are: 66901
Use id=% instead of esmiles to print other entries.
mformula = Cl1
iupac = chlorane doublet radical
PubChem = 313
PubChem LCSS = 313
cas = 7647-01-0
kegg = C01327 D02057
synonyms = hydrochloric acid; hydrogen chloride; 7647-01-0; Muriatic acid; chlorane; Chlorohydric acid; Acide chlorhydrique; Chlorwasserstoff; Spirits of salt; Hydrogen chloride (HCl); Anhydrous hydrochloric acid; Chloorwaterstof; Chlorowodor; Acido cloridrico; Aqueous hydrogen chloride; chlorure d'hydrogene; Hydrochloric acid gas; Marine acid; monohydrochloride; Spirit of salt; UNII-QTT17582CB; NSC 77365; HCl; CHEBI:17883; Hydrogen chloride (acid); [HCl]; MFCD00011324; QTT17582CB; E507; Bowl Cleaner; Hydrogen chloride, 1M solution in ethyl acetate; 4-D Bowl Sanitizer; Chlorowodor [Polish]; Hydrochloric Acid Solution, 1N; Emulsion Bowl Cleaner; Caswell No. 486; Hydrogenchlorid; Chloorwaterstof [Dutch]; o-Tolidine Dihydrochloride Solution; Hydrochloric acid [JAN]; Chlorwasserstoff [German]; Hydrogen Chloride - Methanol Reagent; Titanium, Reference Standard Solution; Vanadium, Reference Standard Solution; Acido clorhidrico; Muriaticum acidum; UN 1789 (solution); Hydrochloric acid, ACS reagent, 37%; UN 1050 (anhydrous); mono hydrochloride; Acido cloridrico [Italian]; Platinum Cobalt Color Standard Solution; White Emulsion Bowl Cleaner; Acido clorhidrico [Spanish]; Varley Poly-Pak Bowl Creme; Acide chlorhydrique [French]; Hydrogen chloride (gas only); Hydrochloric Acid Solution, 0.2N (N/5); Hydrochloric Acid Solution, 0.5N (N/2); Chlorure d'hydrogene [French]; Hydrochloric acid, 0.1 N standard solution; Chloruro de hidrogeno; HSDB 545; Hydrochloric Acid Solution, 0.1N (N/10); Chloruro de hidrogeno [Spanish]; Hygeia Creme Magic Bowl Cleaner; Percleen Bowl and Urinal Cleaner; Hydrochloric acid, pure, 25% solution in water; Hydrogen chloride, ca. 0.5M solution in methanol; EINECS 231-595-7; UN1050; UN1789; UN2186; Anhydrous hydrogen chloride; Hydrochloric acid, 4N solution in 1,4-Dioxane/water; Wuest Bowl Cleaner Super Concentrated; Chlorure d'hydrogene anhydre [French]; Cloruro de hidrogeno anhidro [Spanish]; EPA Pesticide Chemical Code 045901; Hydrochloric acid, for analysis, 25% solution in water; Hydrochloric acid, pure, fuming, 37% solution in water; Chlorure d'hydrogene anhydre; Cloruro de hidrogeno anhidro; UN 2186 (refrigerated liquefied gas); chloro; chlorum; hydochloride; hydrochlorie; hydrochoride; hydrocloride; Hydrochloric acid, ACS reagent, ca. 37% solution in water; Hydrogen chloride, 1.25M solution in ethanol, AcroSeal(R); Hydrogen chloride, pure, 5 to 6N solution in 2-propanol; Salzsaeure; Hydrochloric acid [JAN:NF]; chloridohydrogen; hydro chloride; hydro-chloride; Hydrochloric acid, for analysis, fuming, 37% solution in water; Hydrogen chloride, 4N solution in 1,4-dioxane, AcroSeal(R); Hydrogen chloride, technical, 4 to 6N solution in 2-propanol; hydrogenchloride; Chloro radical; Soldering acid; chlorhydric acid; hydochloric acid; hydogen chloride; hydrochoric acid; hydrocloric acid; hydrogen chlorid; hydrogen choride; hydrogen cloride; hyrochloric acid; hyrogen chloride; Liriopesides-B; Hydrogen chloride, 5 to 6N solution in 2-propanol, AcroSeal(R); Hydrogen chloride, pure, 1N solution in diethyl ether, AcroSeal(R); Hydrogen chloride, pure, 2N solution in diethyl ether, AcroSeal(R); hvdrochloric acid; hvdrogen chloride; hydorchloric acid; hydrochioric acid; hydrochloric aicd; hydrochloric-acid; hydrogen-chloride; hyrdochloric acid; Hydrochloric ccid; Spirits of salts; Wasserstoffchlorid; monohydro-chloride; Sibiricose-A6; hydrogen ch1oride; hydro chloric acid; hydro-chloric acid; hydrochloric ac id; Varley's Ocean Blue Scented Toilet Bowl Cleaner; cloruro de hidrogeno; hydro- chloric acid; Hydrogen chloride - methanol solution; N-s/-Boc-D-lysine; H-Cl; Hydrochloric acid 37%; HCL]; Hydrochloric acid, 37%; Hydrogen chloric anhydrous; Now South Safti-Sol Brand Concentrated Bowl Cleanse with Magic Actio; Hydrochloric acid 36% by weight or more HCl; 17Cl; EC 231-595-7; Hydrochloric acid, solution; Hydrochloric acid, anhydrous; Hydrogen chloride, anhydrous; Hydrogen-chloride-anhydrous-; hydrochloric acid for technical; Hydrochloric acid (JP15/NF); Hydrochloric Acid Solution, 2N; CHEMBL1231821; DTXSID2020711; Hydrochloric acid (JP17/USP); CHEBI:23116; Hydrochloric acid solution, 1 M; Hydrochloric acid solution, 2 M; Hydrochloric acid solution, 6 M; hydrogen chloride ethanol solution; Hydrogen chloride, 4M in dioxane; DTXSID801014230; Hydrochloric Acid Concentrate, 1N; Hydrochloric acid solution, 12 M; Hydrochloric Acid Solution, 0.1N; Hydrochloric acid, AR, 35-37%; Hydrochloric acid, LR, 35-38%; Hydrochloric Acid Concentrate, 10N; Hydrochloric acid solution, 0.1 M; Hydrochloric acid solution, 0.1 N; Hydrochloric acid solution, 0.2 M; Hydrochloric acid solution, 0.5 M; Hydrochloric acid solution, 1.0 N; Hydrochloric acid, 36%, technical; NSC77365; Hydrogen Chloride - Butanol Reagent; SASRIN resin (200-400 mesh); BDBM50499188; Hydrochloric acid solution, 0.01 M; Hydrochloric acid solution, 0.02 M; Hydrochloric acid solution, 0.05 M; Hydrochloric acid, 37%, extra pure; NSC-77365; STL282413; Hydrochloric acid, p.a., 31-33%; Hydrogen chloride, 1M in acetic acid; Hydrogen chloride, refrigerated liquid; AKOS015843726; CCG-221928; DB13366; Hydrochloric acid, puriss., 30-33%; Hydrochloric acid, reagent grade, 37%; Hydrogen chloride, 1M in diethyl ether; Hydrogen chloride, 2M in diethyl ether; MCULE-7728164114; NA 1789; UN 1050; UN 1789; UN 2186; Hydrochloric acid, 1N standard solution; Hydrogen chloride, puriss., >=99.7%; Hydrogen chloride, puriss., >=99.8%; Hydrochloric acid ACS grade 36.5-38%; Hydrochloric acid, technical grade, 30%; 2647-01-0; Hydrochloric acid (acid aerosols including mists, vapors, gas, fog, and other airborne forms of any particle size); H-Lys(2,4-dichloro-Z)-OBzl (c){ HCl; Hydrochloric acid solution, puriss., 36%; 1N Hydrochloric Acid aqueous (+/-0.1N); 3N Hydrochloric Acid aqueous (+/-0.2N); 5N Hydrochloric Acid aqueous (+/-0.2N); DS-002721; Hydrochloric acid, 5% v/v aqueous solution; Hydrochloric acid, puriss. p.a., >=32%; Hydrogen chloride solution 1.6M in ethanol; Hydrogen chloride solution 3.3M in ethanol; Hydrogen chloride solution 4.0M in dioxane; Hydrogen chloride, 25% (w/w) in Methanol; FT-0627124; FT-0628063; FT-0699355; FT-0699890; FT-0699899; FT-0700010; H1060; H1062; H1202; H1203; H1277; Hydrochloric acid, 10% v/v aqueous solution; Hydrochloric acid, 50% v/v aqueous solution; Hydrochloric acid, puriss., 24.5-26.0%; Hydrochloric acid, puriss., 37.0-38.0%; Hydrogen chloride solution 1.25M in ethanol; Hydrogen chloride solution 3.0M in Methanol; Hydrogen chloride solution, 3M in 1-Butanol; Hydrogen chloride, ReagentPlus(R), >=99%; Hydrochloric Acid Solution, 0.02N (N/50); Hydrogen chloride solution, 4.0 M in dioxane; Hydrogen chloride, nominally 2.5M in ethanol; C01327; D02057; Hydrochloric Acid Solution, 0.01N (N/100); Hydrochloric acid, 0.01N Standardized Solution; Hydrochloric acid, 0.05N Standardized Solution; Hydrochloric acid, 0.1N Standardized Solution; Hydrochloric acid, 0.5N Standardized Solution; Hydrochloric acid, 1.0N Standardized Solution; Hydrochloric acid, 5.0N Standardized Solution; Hydrochloric acid, 6.0N Standardized Solution; Hydrogen chloride - Butanol Reagent (5-10%); Hydrogen chloride solution 0.1M in isopropanol; Hydrogen chloride solution 1.0M in acetic acid; Hydrogen chloride - Methanol Reagent (5-10%); Hydrogen chloride solution 1.0M in ethyl acetate; Hydrogen chloride solution 1.25M in isopropanol; Hydrogen chloride, ca 0.5M solution in methanol; 4-methyl-pyridine-2-carboxylic acid hydrochloride; Hydrochloric acid solution, puriss. p.a., >=25%; Hydrogen chloride solution 1.0 M in diethyl ether; Hydrogen chloride solution 2.0 M in diethyl ether; Hydrogen chloride solution, 1.0 M in acetic acid; Hydrogen chloride, 3M in cyclopentyl methyl ether; Q211086; Hydrochloric acid solution, 32 wt. % in H2O, FCC; Hydrochloric acid, pure, ca. 32% solution in water; Hydrochloric acid, SAJ first grade, 35.0-37.0%; Hydrochloric acid, solution [UN1789] [Corrosive]; Hydrogen chloride - ethanol solution, 3% in ethanol; Hydrogen chloride anhydrous [UN1050] [Poison gas]; Hydrogen chloride solution 3.95M-4.40M in dioxane; Hydrogen chloride solution 5.0-6.0M in isopropanol; Hydrogen chloride solution, 1.0 M in diethyl ether; Hydrogen chloride solution, 1M in isopropyl acetate; Hydrogen chloride solution, 2.0 M in diethyl ether; J-006148; Hydrochloric acid solution, BioXtra, ~0.1 M in H2O; Hydrochloric acid, Environmental Grade Plus, 33-36%; Hydrochloric acid, JIS special grade, 35.0-37.0%; Hydrochloric acid, p.a., ACS reagent, 36.5-38.0%; Hydrochloric acid, SAJ super special grade, >=35.0%; Hydrogen chloride, 3M solution in CPME, AcroSeal(R); Hydrogen chloride, anhydrous [UN1050] [Poison gas]; Hydrogen chloride - ethanol solution, 0.1 M in ethanol; Hydrochloric acid solution, protein sequencing grade, liquid; Hydrochloric acid solution, SAJ first grade, 9.5-10.0%; Hydrochloric acid, for analysis, ca. 32% solution in water; Hydrochloric acid, purum p.a., fuming 37%, >=37% (T); Hydrogen chloride, 1M solution in acetic acid, AcroSeal(R); Hydrogen chloride, 5 to 6M solution in 2-propanol, pure; Hydrochloric acid solution, 0.1 M, for 3S adapter technology; Hydrochloric acid, 37%, for analysis, (max. 0.000001% Hg); Hydrogen chloride, 1M solution in ethyl acetate, AcroSeal(R); Hydrogen chloride, ca. 0.5M solution in methanol, AcroSeal(R); Hydrogen chloride, refrigerated liquid [UN2186] [Poison gas]; Hydrochloric acid solution, ~6 M in H2O, for amino acid analysis; Hydrochloric acid, 36.5-38.0%, BioReagent, for molecular biology; Hydrochloric acid, 37 wt. % in H2O, 99.999% trace metals basis; Hydrochloric acid, BioUltra, >=32% (T), suitable for luminescence; Hydrochloric acid, suitable for amino acid analysis, 35.0-37.0%; Hydrochloric acid, suitable for arsenic determination, 35.0-37.0%; Hydrogen chloride solution 3.0M in cyclopentyl methyl ether (CPME); Hydrogen chloride solution, 3 M in cyclopentyl methyl ether (CPME); Hydrogen chloride solution, 3 M in methanol, for GC derivatization; Hydrochloric acid solution, 1.0 N, BioReagent, suitable for cell culture; Hydrochloric acid solution, volumetric, 0.1 M HCl (0.1N), endotoxin free; Hydrochloric acid, suitable for determination of toxic metals, >=35.0%; Hydrogen chloride - ethanol solution, ~1.25 M HCl, for GC derivatization; Hydrogen chloride - methanol solution, ~1.25 M HCl, for GC derivatization; Hydrogen chloride solution, 0.5 M in methanol, for GC derivatization; Hydrochloric acid ACS grade 36.5-38% for Biochemistry and Molecular biology; Hydrochloric acid, meets analytical specification of Ph. Eur., BP, NF, fuming, 36.5-38%; Hydrochloric acid, p.a., ACS reagent, reag. ISO, reag. Ph. Eur., 37.0-38.0%; Hydrochloric acid, semiconductor grade PURANAL(TM) (Honeywell 17823), fuming 37%, 37-38%; Hydrochloric acid, semiconductor grade PURANAL(TM) (Honeywell 17863), >=32%; Hydrochloric acid, semiconductor grade SLSI PURANAL(TM) (Honeywell 17302), fuming 37%; Hydrochloric acid, semiconductor grade VLSI PURANAL(TM) (Honeywell 17610), fuming 37%; Hydrogen chloride - 1-butanol solution, ~3 M in 1-butanol, for GC derivatization; Hydrogen chloride - 2-propanol solution, ~1.25 M HCl (T), for GC derivatization; 185912-82-7; Chloride atomic spectroscopy standard concentrate 10.00 g Cl-, 10.00 g/L, for 1 l standard solution, analytical standard; Hydrochloric acid concentrate, 0.1N, Dissolution Media Concentrate, Dilute to 25L to conform to USP & EP; Hydrochloric acid concentrate, 0.1N, Dissolution Media Concentrate, Dilute to 6L to conform to USP & EP; Hydrochloric acid, puriss. p.a., ACS reagent, reag. ISO, reag. Ph. Eur., fuming, >=37%, APHA: <=10
Search Links to Other Online Resources (may not be available):
- Google Structure Search
- EPA CompTox Database
- Chemical Entities of Biological Interest (ChEBI)
- NIH ChemIDplus - A TOXNET DATABASE
- The Human Metabolome Database (HMDB)
- OECD eChemPortal
- Google Scholar
+==================================================+
|| Molecular Calculation ||
+==================================================+
Id = 66901
NWOutput = Link to NWChem Output (download)
Datafiles:
density.cube-973125-2021-11-21-9:54:14 (download)
homo-alpha.cube-973125-2021-11-21-9:54:14 (download)
homo-beta.cube-973125-2021-11-21-9:54:14 (download)
lumo-alpha.cube-973125-2021-11-21-9:54:14 (download)
lumo-beta.cube-973125-2021-11-21-9:54:14 (download)
image_resset: api/image_reset/66901
Calculation performed by Eric Bylaska - constance.pnl.gov
Numbers of cpus used for calculation = 48
Calculation walltime = 229.300000 seconds (0 days 0 hours 3 minutes 49 seconds)
+----------------+
| Energetic Data |
+----------------+
Id = 66901
iupac = chlorane doublet radical
mformula = Cl1
inchi = InChI=1S/Cl
inchikey = ZAMOUSCENKQFHK-UHFFFAOYSA-N
esmiles = [Cl] theory{pspw} xc{b3lyp} basis{100.0 Ry} solvation_type{None} ^{0} mult{2}
calculation_type = ov
theory = pspw
xc = b3lyp
basis = 100.0 Ry
charge,mult = 0 2
energy = -15.010929 Hartrees
enthalpy correct.= 0.002359 Hartrees
entropy = 36.570 cal/mol-K
solvation energy = 0.000 kcal/mol solvation_type = None
Sitkoff cavity dispersion = 1.483 kcal/mol
Honig cavity dispersion = 3.117 kcal/mol
ASA solvent accesible surface area = 124.690 Angstrom2
ASA solvent accesible volume = 130.924 Angstrom3
+-----------------+
| Structural Data |
+-----------------+
JSmol: an open-source HTML5 viewer for chemical structures in 3D
Number of Atoms = 1
Units are Angstrom for bonds and degrees for angles
Type I J K L M Value
----------- ----- ----- ----- ----- ----- ----------
+-----------------+
| Eigenvalue Data |
+-----------------+
Id = 66901
iupac = chlorane doublet radical
mformula = Cl1
InChI = InChI=1S/Cl
smiles = [Cl]
esmiles = [Cl] theory{pspw} xc{b3lyp} basis{100.0 Ry} solvation_type{None} ^{0} mult{2}
theory = pspw
xc = b3lyp
basis = 100.0 Ry
charge = 0
mult = 2
solvation_type = None
twirl webpage = TwirlMol Link
image webpage = GIF Image Link
Unrestricted Eigevalue Spectra
alpha beta
- - - - -- 1.10 eV - - - - -- 1.10 eV
---- ---- ----------
----------LUMO= -0.29 eV ----------
----------LUMO= -6.49 eV
HOMO= -9.14 eV++++ ++++
HOMO= -9.53 eV++++ ++++
++++++++++
-21.55 eV++++++++++
-23.09 eV++++++++++
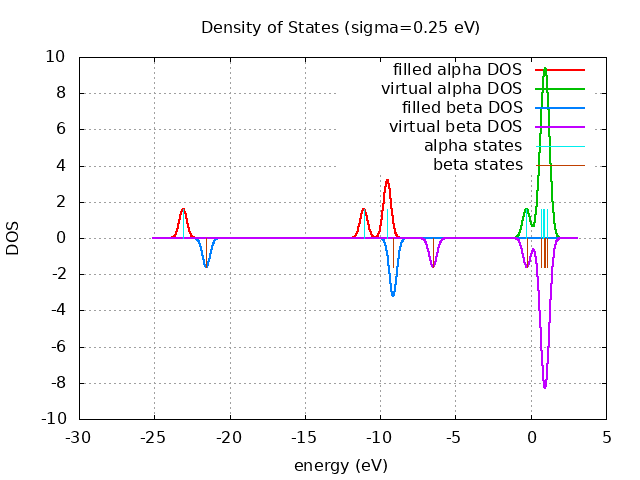
spin eig occ ---------------------------- alpha -9.53 1.00 alpha -9.53 1.00 alpha -11.09 1.00 alpha -23.09 1.00 alpha 1.10 0.00 alpha 1.09 0.00 alpha 1.09 0.00 alpha 0.89 0.00 alpha 0.89 0.00 alpha 0.84 0.00 alpha 0.65 0.00 alpha -0.29 0.00 beta -9.14 1.00 beta -9.14 1.00 beta -21.55 1.00 beta 1.10 0.00 beta 1.10 0.00 beta 0.93 0.00 beta 0.90 0.00 beta 0.90 0.00 beta 0.68 0.00 beta -0.26 0.00 beta -6.49 0.00
+----------------------------------------+ | Vibrational Density of States Analysis | +----------------------------------------+ Total number of frequencies = 3 Total number of negative frequencies = 0 Number of lowest frequencies = 0 (frequency threshold = 500 ) Exact dos norm = -3.000 Generating vibrational DOS Generating model vibrational DOS to have a proper norm 10.00 0.00 0.00 0.00 50.00 0.00 0.00 0.00 100.00 0.00 0.00 0.00 Writing vibrational density of states (DOS) to vdos.dat Writing model vibrational density of states (DOS_FIXED) to vdos-model.dat Temperature= 298.15 zero-point correction to energy = 0.000 kcal/mol ( 0.000000) vibrational contribution to enthalpy correction = 0.000 kcal/mol ( 0.000000) vibrational contribution to Entropy = 0.000 cal/mol-k hindered rotor enthalpy correction = 0.000 kcal/mol ( 0.000000) hindered rotor entropy correction = 0.000 cal/mol-k
DOS sigma = 10.000000
- vibrational DOS enthalpy correction = 0.000000 kcal/mol ( 0.000 kcal/mol)
- model vibrational DOS enthalpy correction = 0.000000 kcal/mol ( 0.000 kcal/mol)
- vibrational DOS Entropy = 0.000000 ( 0.000 cal/mol-k)
- model vibrational DOS Entropy = 0.000000 ( 0.000 cal/mol-k)
- original gas Energy = -15.010929 (-9419.500 kcal/mol)
- original gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- model DOS gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- original gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- model DOS gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- original gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- model DOS gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- original sol Free Energy = -15.025946 (-9428.923 kcal/mol)
- unadjusted DOS sol Free Energy = -15.025946 (-9428.923 kcal/mol)
- model DOS sol Free Energy = -15.025946 (-9428.923 kcal/mol)
DOS sigma = 50.000000
- vibrational DOS enthalpy correction = 0.000000 kcal/mol ( 0.000 kcal/mol)
- model vibrational DOS enthalpy correction = 0.000000 kcal/mol ( 0.000 kcal/mol)
- vibrational DOS Entropy = 0.000000 ( 0.000 cal/mol-k)
- model vibrational DOS Entropy = 0.000000 ( 0.000 cal/mol-k)
- original gas Energy = -15.010929 (-9419.500 kcal/mol)
- original gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- model DOS gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- original gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- model DOS gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- original gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- model DOS gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- original sol Free Energy = -15.025946 (-9428.923 kcal/mol)
- unadjusted DOS sol Free Energy = -15.025946 (-9428.923 kcal/mol)
- model DOS sol Free Energy = -15.025946 (-9428.923 kcal/mol)
DOS sigma = 100.000000
- vibrational DOS enthalpy correction = 0.000000 kcal/mol ( 0.000 kcal/mol)
- model vibrational DOS enthalpy correction = 0.000000 kcal/mol ( 0.000 kcal/mol)
- vibrational DOS Entropy = 0.000000 ( 0.000 cal/mol-k)
- model vibrational DOS Entropy = 0.000000 ( 0.000 cal/mol-k)
- original gas Energy = -15.010929 (-9419.500 kcal/mol)
- original gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- model DOS gas Enthalpy = -15.008570 (-9418.020 kcal/mol, delta= 0.000)
- original gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- model DOS gas Entropy = 0.000058 ( 36.570 cal/mol-k,delta= 0.000)
- original gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- model DOS gas Free Energy = -15.025946 (-9428.923 kcal/mol, delta= 0.000)
- original sol Free Energy = -15.025946 (-9428.923 kcal/mol)
- unadjusted DOS sol Free Energy = -15.025946 (-9428.923 kcal/mol)
- model DOS sol Free Energy = -15.025946 (-9428.923 kcal/mol)
Normal Mode Frequency (cm-1) IR Intensity (arbitrary)
----------- ---------------- ------------------------
1 0.000 0.000
2 0.000 0.000
3 0.000 0.000
No Hindered Rotor Data
+-------------------------------------+
| Reactions Contained in the Database |
+-------------------------------------+
Reactions Containing INCHIKEY = ZAMOUSCENKQFHK-UHFFFAOYSA-N
Reactionid Erxn(gas) Hrxn(gas) Grxn(gas) delta_Solv Grxn(aq) ReactionType Reaction
20298 6.560 8.305 9.933 32.484 42.416 AB + C --> AC + B "C(=S)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> S=[CH-]O xc{m06-2x} + [Cl] xc{m06-2x}"
20273 25.776 27.902 30.014 61.256 91.270 AB + C --> AC + B "C(=O)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> O[CH-][O] xc{m06-2x} + [Cl] xc{m06-2x}"
19581 -68.007 -65.150 -62.915 15.081 -47.834 AB + C --> AC + B "[N]CC(C[N])=Cc(cccc1)c1Cl + hydroxide ^{-1} --> [N]CC(=Cc1ccccc1O)C[N] + [Cl] ^{-1}"
19408 89.757 86.460 78.400 1.294 79.694 AB --> A + B "CCl xc{pbe} --> [CH3] xc{pbe} + [Cl] xc{pbe}"
19407 89.757 86.460 78.400 1.294 79.694 AB --> A + B "CCl xc{pbe} --> [CH3] xc{pbe} + [Cl] xc{pbe}"
19396 -241.520 -234.305 -231.635 31.503 -2.932 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> CCl + [Cl] ^{-2} mult{2}"
19395 -268.394 -264.957 -269.066 158.435 86.569 AB + C --> AC + B "ClC(Cl)Cl ^{1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> ClCCl ^{-1} mult{2} + [Cl] ^{1}"
19333 -8.615 -9.624 -14.807 -77.587 6.207 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
19332 -8.615 -9.624 -14.807 -77.587 6.207 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
19331 7.259 6.837 1.624 -16.540 -14.916 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
19330 7.259 6.837 1.624 -16.540 -14.916 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
19329 129.646 134.661 140.040 -132.810 7.230 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [OH-] ^{-1} --> OCCl + [Cl] ^{-2} mult{2}"
19328 167.244 170.768 175.471 -154.768 20.702 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [SH-] ^{-1} --> SCCl + [Cl] ^{-2} mult{2}"
19268 -62.103 -59.244 -57.690 12.890 -44.800 AB + C --> AC + B "TCE + hydroxide ^{-1} --> OC=C(Cl)Cl + [Cl] ^{-1}"
18742 89.757 86.459 78.401 1.334 79.734 AB --> A + B "CCl xc{pbe} --> [CH3] xc{pbe} + [Cl] xc{pbe}"
18741 89.757 86.459 78.401 1.334 79.734 AB --> A + B "CCl xc{pbe} --> [CH3] xc{pbe} + [Cl] xc{pbe}"
18212 9.722 11.012 14.584 -15.579 -0.995 AC + BD --> A + B + CD "C(Cl)(Cl)(Cl)Cl + nitrate ^{-1} --> ClC([O]=[N](=O)=O)(Cl)Cl + [Cl] ^{-1}"
18185 367.324 365.501 358.218 -161.709 196.510 AB --> A + B "TCE --> ClC(=[CH])Cl ^{-1} + [Cl] ^{1}"
18184 367.324 365.501 358.218 -161.709 196.510 AB --> A + B "TCE --> ClC(=[CH])Cl ^{-1} + [Cl] ^{1}"
18171 5.499 3.436 -5.273 -79.782 13.545 AB --> A + B "TCE + [SHE] --> ClC(=[CH])Cl mult{2} + [Cl] ^{-1}"
18170 5.499 3.436 -5.273 -79.782 13.545 AB --> A + B "TCE + [SHE] --> ClC(=[CH])Cl mult{2} + [Cl] ^{-1}"
17322 343.697 345.058 346.698 -124.920 221.778 AB + C --> AC + B "Cl/C=C/Cl + [OH] mult{2} --> OC=CCl ^{-1} mult{2} + [Cl] ^{1}"
17293 129.744 133.747 136.167 -138.305 -2.139 AB + C --> AC + B "[CH]=[Cl] ^{-1} mult{2} + [OH-] ^{-1} --> [CH]O + [Cl] ^{-2} mult{2}"
17279 165.679 169.211 173.843 -154.659 19.183 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [SH-] ^{-1} --> SCCl + [Cl] ^{-2} mult{2}"
17277 158.840 164.381 170.722 -148.874 21.848 AB + C --> AC + B "OC(Cl)(Cl)Cl ^{-1} mult{2} + [OH-] ^{-1} --> OC(Cl)(Cl)O + [Cl] ^{-2} mult{2}"
17276 170.107 169.191 160.499 -29.522 130.977 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{1}"
17275 170.107 169.191 160.499 -29.522 130.977 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{1}"
17274 -237.308 -231.418 -232.490 182.498 147.208 AB + C --> AC + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> CCl ^{-1} mult{2} + [Cl] ^{1}"
17269 154.233 152.730 144.068 -90.568 152.100 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{1}"
17268 154.233 152.730 144.068 -90.568 152.100 AB --> A + B "[Cl]=[CH2]=[Cl] ^{1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{1}"
17265 402.184 399.685 391.287 -159.590 231.697 AB --> A + B "Cl[CH]Cl mult{2} --> [CH]=[Cl] ^{-1} mult{2} + [Cl] ^{1}"
17264 402.184 399.685 391.287 -159.590 231.697 AB --> A + B "Cl[CH]Cl mult{2} --> [CH]=[Cl] ^{-1} mult{2} + [Cl] ^{1}"
17262 76.088 76.943 77.193 10.989 88.183 AB + C --> AC + B "ClC(Cl)Cl ^{1} mult{2} + [OH-] ^{-1} --> OC(Cl)Cl ^{-1} mult{2} + [Cl] ^{1}"
17261 130.473 128.499 119.642 -94.452 123.790 AB --> A + B "ClC(Cl)Cl ^{1} mult{2} + [SHE] --> Cl[CH]Cl ^{-1} + [Cl] ^{1}"
17260 130.473 128.499 119.642 -94.452 123.790 AB --> A + B "ClC(Cl)Cl ^{1} mult{2} + [SHE] --> Cl[CH]Cl ^{-1} + [Cl] ^{1}"
17253 -243.085 -235.862 -233.263 31.612 -4.451 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> CCl + [Cl] ^{-2} mult{2}"
17250 205.448 204.371 196.895 -231.790 63.706 AB --> A + B "Cl[CH]Cl ^{-1} + [SHE] --> [CH]Cl + [Cl] ^{-2} mult{2}"
17249 205.448 204.371 196.895 -231.790 63.706 AB --> A + B "Cl[CH]Cl ^{-1} + [SHE] --> [CH]Cl + [Cl] ^{-2} mult{2}"
17248 -266.828 -263.400 -267.438 158.326 88.088 AB + C --> AC + B "ClC(Cl)Cl ^{1} mult{2} + [H+] ^{1} + 2.00 [SHE] --> ClCCl ^{-1} mult{2} + [Cl] ^{1}"
17247 -10.180 -11.181 -16.435 -77.477 4.688 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
17246 -10.180 -11.181 -16.435 -77.477 4.688 AB --> A + B "ClCCl ^{-1} mult{2} + [SHE] --> [CH2]Cl ^{-1} + [Cl] ^{-1}"
17245 128.080 133.104 138.412 -132.701 5.711 AB + C --> AC + B "ClCCl ^{-1} mult{2} + [OH-] ^{-1} --> OCCl + [Cl] ^{-2} mult{2}"
17237 5.694 5.280 -0.004 -16.431 -16.435 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
17236 5.694 5.280 -0.004 -16.431 -16.435 AB --> A + B "ClCCl ^{-1} mult{2} --> [CH2]Cl mult{2} + [Cl] ^{-1}"
17208 -21.902 -22.965 -31.438 -81.432 -14.271 AB --> A + B "Cl[C](Cl)Cl mult{2} + [SHE] --> Cl[C]Cl + [Cl] ^{-1}"
17207 -21.902 -22.965 -31.438 -81.432 -14.271 AB --> A + B "Cl[C](Cl)Cl mult{2} + [SHE] --> Cl[C]Cl + [Cl] ^{-1}"
17135 164.842 163.502 155.204 -35.499 119.705 AB --> A + B "ClC(Cl)Cl ^{1} mult{2} --> Cl[CH]Cl mult{2} + [Cl] ^{1}"
17134 164.842 163.502 155.204 -35.499 119.705 AB --> A + B "ClC(Cl)Cl ^{1} mult{2} --> Cl[CH]Cl mult{2} + [Cl] ^{1}"
15555 381.572 379.298 370.028 -157.091 212.937 AB --> A + B "chloroform --> Cl[CH]Cl ^{-1} + [Cl] ^{1}"
15554 381.572 379.298 370.028 -157.091 212.937 AB --> A + B "chloroform --> Cl[CH]Cl ^{-1} + [Cl] ^{1}"
15413 409.886 406.586 397.929 -158.712 239.217 AB --> A + B "ClCCl --> [CH2]Cl ^{-1} + [Cl] ^{1}"
15412 409.886 406.586 397.929 -158.712 239.217 AB --> A + B "ClCCl --> [CH2]Cl ^{-1} + [Cl] ^{1}"
15194 5.027 7.653 9.598 0.000 9.598 AB + C --> AC + B "C(=S)Cl theory{pspw4} + [OH-] theory{pspw4} --> S=[CH-]O theory{pspw4} + [Cl] theory{pspw4}"
15132 6.559 8.305 9.932 38.334 48.266 AB + C --> AC + B "C(=S)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> S=[CH-]O xc{m06-2x} + [Cl] xc{m06-2x}"
15131 6.358 7.992 9.589 37.942 47.531 AB + C --> AC + B "C(=S)Cl xc{pbe0} + [OH-] xc{pbe0} --> S=[CH-]O xc{pbe0} + [Cl] xc{pbe0}"
15130 14.001 15.407 16.958 38.250 55.209 AB + C --> AC + B "C(=S)Cl xc{pbe} + [OH-] xc{pbe} --> S=[CH-]O xc{pbe} + [Cl] xc{pbe}"
15129 9.210 10.810 12.379 38.193 50.572 AB + C --> AC + B "C(=S)Cl xc{b3lyp} + [OH-] xc{b3lyp} --> S=[CH-]O xc{b3lyp} + [Cl] xc{b3lyp}"
15084 25.776 27.902 30.014 67.106 97.120 AB + C --> AC + B "C(=O)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> O[CH-][O] xc{m06-2x} + [Cl] xc{m06-2x}"
15083 22.006 24.402 26.524 68.575 95.099 AB + C --> AC + B "C(=O)Cl xc{pbe0} + [OH-] xc{pbe0} --> O[CH-][O] xc{pbe0} + [Cl] xc{pbe0}"
15082 28.328 30.296 32.452 63.614 96.067 AB + C --> AC + B "C(=O)Cl xc{pbe} + [OH-] xc{pbe} --> O[CH-][O] xc{pbe} + [Cl] xc{pbe}"
15081 25.787 28.019 30.168 67.044 97.212 AB + C --> AC + B "C(=O)Cl + [OH-] --> O[CH-][O] + [Cl]"
15005 -0.656 -2.368 -10.403 -82.223 5.974 AB --> A + B "Cl[CH]Cl mult{2} + [SHE] --> [CH]=[Cl] + [Cl] ^{-1}"
15004 -0.656 -2.368 -10.403 -82.223 5.974 AB --> A + B "Cl[CH]Cl mult{2} + [SHE] --> [CH]=[Cl] + [Cl] ^{-1}"
10994 317.730 317.720 312.215 -178.998 133.218 AB --> A + B "ClCl --> [Cl] ^{-1} + [Cl] ^{1}"
9272 55.203 55.182 49.656 0.000 49.656 AB --> A + B "[Cl][Cl] theory{pspw4} xc{b3lyp} --> 2 [Cl] mult{2} theory{pspw4} xc{b3lyp}"
9271 55.139 55.117 49.590 0.000 49.590 AB --> A + B "[Cl][Cl] theory{pspw} xc{b3lyp} --> 2 [Cl] mult{2} theory{pspw} xc{b3lyp}"
9270 55.446 55.433 49.914 0.104 50.019 AB --> A + B "[Cl][Cl] xc{b3lyp} basis{Def2-TZVPD} --> 2 [Cl] mult{2} xc{b3lyp} basis{Def2-TZVPD}"
9239 58.205 58.231 52.756 0.262 53.018 AB --> A + B "[Cl][Cl] xc{blyp} basis{Def2-TZVPD} --> 2 [Cl] mult{2} xc{blyp} basis{Def2-TZVPD}"
9222 59.667 59.615 54.055 0.097 54.152 AB --> A + B "[Cl][Cl] xc{pbe0} basis{Def2-TZVPD} --> 2 [Cl] mult{2} xc{pbe0} basis{Def2-TZVPD}"
9209 65.577 65.566 60.045 0.205 60.250 AB --> A + B "[Cl][Cl] xc{pbe} basis{Def2-TZVPD} --> 2 [Cl] mult{2} xc{pbe} basis{Def2-TZVPD}"
9190 83.021 82.984 77.433 -0.003 77.430 AB --> A + B "[Cl][Cl] xc{lda} basis{Def2-TZVPD} --> 2 [Cl] mult{2} xc{lda} basis{Def2-TZVPD}"
9158 57.933 57.950 52.464 0.000 52.464 AB --> A + B "[Cl][Cl] theory{pspw4} xc{blyp} --> 2 [Cl] mult{2} theory{pspw4} xc{blyp}"
9157 64.738 64.748 59.258 0.000 59.258 AB --> A + B "[Cl][Cl] theory{pspw} xc{blyp} --> 2 [Cl] mult{2} theory{pspw} xc{blyp}"
9156 56.338 56.365 50.904 0.070 50.974 AB --> A + B "[Cl][Cl] xc{blyp} --> 2 [Cl] mult{2} xc{blyp}"
9032 84.150 84.101 78.540 0.000 78.540 AB --> A + B "[Cl][Cl] theory{pspw4} xc{lda} --> 2 [Cl] mult{2} theory{pspw4} xc{lda}"
9031 84.151 84.103 78.542 0.000 78.542 AB --> A + B "[Cl][Cl] theory{pspw} xc{lda} --> 2 [Cl] mult{2} theory{pspw} xc{lda}"
8989 53.230 53.220 47.715 0.162 47.878 AB --> A + B "[Cl][Cl] xc{b3lyp} --> 2 [Cl] mult{2} xc{b3lyp}"
8984 59.436 59.374 53.805 0.000 53.805 AB --> A + B "[Cl][Cl] theory{pspw4} xc{pbe0} --> 2 [Cl] mult{2} theory{pspw4} xc{pbe0}"
8983 59.367 59.305 53.735 0.000 53.735 AB --> A + B "[Cl][Cl] theory{pspw} xc{pbe0} --> 2 [Cl] mult{2} theory{pspw} xc{pbe0}"
8982 57.285 57.235 51.690 0.045 51.735 AB --> A + B "[Cl][Cl] xc{pbe0} --> 2 [Cl] mult{2} xc{pbe0}"
8979 65.075 65.051 59.517 0.000 59.517 AB --> A + B "[Cl][Cl] theory{pspw4} xc{pbe} --> 2 [Cl] mult{2} theory{pspw4} xc{pbe}"
8978 65.077 65.053 59.519 0.000 59.519 AB --> A + B "[Cl][Cl] theory{pspw} xc{pbe} --> 2 [Cl] mult{2} theory{pspw} xc{pbe}"
8977 63.465 63.457 57.951 0.133 58.084 AB --> A + B "[Cl][Cl] xc{pbe} --> 2 [Cl] mult{2} xc{pbe}"
8969 79.987 79.954 74.418 0.145 74.564 AB --> A + B "[Cl][Cl] xc{lda} --> 2 [Cl] mult{2} xc{lda}"
8010 199.671 199.526 191.872 -236.037 54.435 AB --> A + B "Cl[C](Cl)Cl ^{-1} + [SHE] --> Cl[C]Cl + [Cl] ^{-2} mult{2}"
8009 199.671 199.526 191.872 -236.037 54.435 AB --> A + B "Cl[C](Cl)Cl ^{-1} + [SHE] --> Cl[C]Cl + [Cl] ^{-2} mult{2}"
7969 371.510 369.871 360.737 -157.131 203.606 AB --> A + B "Cl[C](Cl)Cl mult{2} --> Cl[C]Cl ^{-1} mult{2} + [Cl] ^{1}"
7968 371.510 369.871 360.737 -157.131 203.606 AB --> A + B "Cl[C](Cl)Cl mult{2} --> Cl[C]Cl ^{-1} mult{2} + [Cl] ^{1}"
7812 7.048 8.340 11.852 -13.138 -1.286 AC + BD --> A + B + CD "C(Cl)(Cl)(Cl)Cl + nitrate ^{-1} --> ClC([O]=[N](=O)=O)(Cl)Cl + [Cl] ^{-1}"
7260 -10.622 -7.855 -6.152 0.871 -5.281 AB + C --> AC + B "CCl + [OH] --> CO + [Cl]"
6199 -71.515 -68.631 -66.873 14.912 -51.961 AB + C --> AC + B "CC(Cl)(Cl)Cl + [OH] ^{-1} --> CC(Cl)(Cl)O + [Cl] ^{-1}"
5997 431.104 426.548 417.695 -152.936 264.759 AB --> A + B "ClC(C)(C)C --> C[C](C)C ^{-1} + [Cl] ^{1}"
5996 431.104 426.548 417.695 -152.936 264.759 AB --> A + B "ClC(C)(C)C --> C[C](C)C ^{-1} + [Cl] ^{1}"
5672 384.139 381.878 372.608 -157.061 215.547 AB --> A + B "chloroform --> Cl[CH]Cl ^{-1} + [Cl] ^{1}"
5671 384.139 381.878 372.608 -157.061 215.547 AB --> A + B "chloroform --> Cl[CH]Cl ^{-1} + [Cl] ^{1}"
5151 360.129 358.555 348.761 -152.440 196.321 AB --> A + B "C(Cl)(Cl)(Cl)Cl --> Cl[C](Cl)Cl ^{-1} + [Cl] ^{1}"
5150 360.129 358.555 348.761 -152.440 196.321 AB --> A + B "C(Cl)(Cl)(Cl)Cl --> Cl[C](Cl)Cl ^{-1} + [Cl] ^{1}"
4807 429.578 430.466 425.044 -119.332 305.712 AB --> A + B "[Na]Cl --> [Na] ^{-1} + [Cl] ^{1}"
4806 429.578 430.466 425.044 -119.332 305.712 AB --> A + B "[Na]Cl --> [Na] ^{-1} + [Cl] ^{1}"
3146 207.748 207.178 205.193 -196.087 9.107 AB --> A + B "Cl[Al](Cl)Cl --> Cl[Al]Cl ^{1} + [Cl] ^{-1}"
2600 227.114 226.380 222.301 -165.777 56.524 AB --> A + B "Cl[Fe](Cl)Cl mult{6} --> Cl[Fe]Cl ^{1} mult{6} + [Cl] ^{-1}"
2408 89.757 86.458 78.399 1.314 79.713 AB --> A + B "CCl xc{pbe} --> [CH3] xc{pbe} + [Cl] xc{pbe}"
2291 87.501 83.978 75.969 0.000 75.969 AB --> A + B "CCl theory{pspw4} --> [CH3] theory{pspw4} + [Cl] theory{pspw4}"
2249 87.544 83.967 75.941 0.000 75.941 AB --> A + B "CCl theory{pspw} --> [CH3] theory{pspw} + [Cl] theory{pspw}"
2248 87.544 83.967 75.941 0.000 75.941 AB --> A + B "CCl theory{pspw} --> [CH3] theory{pspw} + [Cl] theory{pspw}"
2243 659.767 659.704 654.192 -641.397 12.795 AB --> A + B "[Al]Cl [Al]Cl ^{2} --> [Al] [Al] ^{3} + [Cl] [Cl] ^{-1}"
2187 82.365 78.876 70.834 1.473 72.307 AB --> A + B "CCl --> [CH3] + [Cl]"
2182 711.352 712.240 706.525 -472.573 233.951 AB --> A + B "Cl[Fe] ^{2} mult{6} --> [Fe] ^{3} mult{6} + [Cl] ^{-1}"
2130 410.781 410.390 398.030 -389.947 8.084 AB --> A + B "Cl[Al]Cl ^{1} --> [Al]Cl ^{2} + [Cl] ^{-1}"
1807 174.076 172.616 164.282 -145.604 18.678 AB + C --> AC + B "ClCC(Cl)CCl --> ClCC1C[Cl]1 ^{+1} + [Cl] ^{-1}"
1806 174.076 172.616 164.282 -145.604 18.678 AB + C --> AC + B "ClCC(Cl)CCl --> ClCC1C[Cl]1 ^{+1} + [Cl] ^{-1}"
1554 -10.622 -7.855 -6.152 0.931 -5.221 AB + C --> AC + B "CCl + [OH] --> CO + [Cl]"
1379 -63.181 -60.726 -59.814 14.191 -45.624 AB + C --> AC + B "Mitotane + hydroxide ^{-1} --> ClC(C(c1ccccc1Cl)c1ccc(cc1)O)Cl + [Cl] ^{-1}"
1336 -63.441 -60.790 -59.600 16.037 -43.564 AB + C --> AC + B "Mitotane + hydroxide ^{-1} --> ClC(C(c1ccccc1O)C1=CC=[C](=[Cl])C=C1)Cl + [Cl] ^{-1}"
1291 -426.506 -421.296 -422.326 178.582 -46.544 AB + C --> AC + B "DDT + [H+] ^{1} + 2.00 [SHE] --> Clc1ccc(cc1)C(C(Cl)(Cl)Cl)c1ccccc1 + [Cl] ^{-1}"
1147 -56.124 -53.531 -51.678 16.036 -35.641 AB + C --> AC + B "C(N)Cl + [OH] ^{-1} --> OCN + [Cl] ^{-1}"
1110 -10.243 -10.090 -10.031 0.000 -10.031 AB + C --> AC + B "C(Br)Br theory{pspw4} + [Cl] ^{-1} theory{pspw4} --> ClCBr theory{pspw4} + [Br] ^{-1} theory{pspw4}"
1039 -49.057 -49.352 -59.438 11.760 -47.677 ABCD + E --> A + BC + DE "ClCC(CCl)Cl + [OH] ^{-1} --> ClC/C=C\Cl + O + [Cl] ^{-1}"
1032 -49.125 -49.523 -59.571 11.278 -48.293 ABCD + E --> A + BC + DE "ClCC(Cl)CCl + [OH-] ^{-1} --> Cl[CH][CH]CCl + [Cl] ^{-1} + O"
780 -50.896 -48.714 -46.818 0.000 -46.818 AB + C --> AC + B "[SiH3]Cl theory{pspw4} + [OH] ^{-1} theory{pspw4} --> O[SiH3] theory{pspw4} + [Cl] ^{-1} theory{pspw4}"
768 227.114 226.380 222.301 -165.777 56.524 AB --> A + B "Cl[Fe](Cl)Cl mult{6} --> Cl[Fe]Cl ^{1} mult{6} + [Cl] ^{-1}"
767 711.352 712.240 706.525 -472.573 233.951 AB --> A + B "Cl[Fe] ^{2} mult{6} --> [Fe] ^{3} mult{6} + [Cl] ^{-1}"
765 207.748 207.178 205.193 -196.087 9.107 AB --> A + B "Cl[Al](Cl)Cl --> Cl[Al]Cl ^{1} + [Cl] ^{-1}"
764 410.781 410.390 398.030 -389.947 8.084 AB --> A + B "Cl[Al]Cl ^{1} --> [Al]Cl ^{2} + [Cl] ^{-1}"
763 659.767 659.704 654.192 -641.397 12.795 AB --> A + B "[Al]Cl xyzdata{Al -4.04269 -0.04643 0.07344 | Cl -6.25346 -0.16386 0.07347} ^{2} --> [Al] xyzdata{Al 3.18558 -0.53312 -0.07187} ^{3} + [Cl] xyzdata{Cl 4.73576 3.37508 0.16662} ^{-1}"
749 -3.693 -4.543 -12.921 -80.119 -93.041 AB --> A + B "Cl[C](Cl)Cl xyzdata{C -4.00022 0.58207 0.96997 | Cl -2.30229 0.97442 1.34210 | Cl -4.34205 -1.12742 1.31115 | Cl -4.38438 0.98070 -0.71795} --> [Cl] xyzdata{Cl 2.75521 1.83484 -0.24354} ^{-1} + Cl[C]Cl xyzdata{C 3.04352 -2.52273 -0.25387 | Cl 1.77592 -1.56644 -1.04794 | Cl 3.94514 -1.50287 0.88596} mult{3}"
699 87.501 83.978 75.969 0.000 75.969 AB --> A + B "CCl theory{pspw4} --> [CH3] theory{pspw4} + [Cl] theory{pspw4}"
698 89.757 86.458 78.399 1.314 79.713 AB --> A + B "CCl xc{pbe} --> [CH3] xc{pbe} + [Cl] xc{pbe}"
697 87.544 83.952 75.924 0.000 75.924 AB --> A + B "CCl theory{pspw} --> [CH3] theory{pspw} + [Cl] theory{pspw}"
696 82.365 78.876 70.835 1.482 72.316 AB --> A + B "CCl --> [CH3] + [Cl]"
493 -54.477 -51.461 -49.334 0.000 -49.334 AB + C --> AC + B "Oc1cccc(Cl)c1 theory{pspw4} + [OH-] ^{-1} theory{pspw4} --> Oc1cccc(c1)O theory{pspw4} + [Cl] ^{-1} theory{pspw4}"
491 -63.344 -60.736 -58.893 13.988 -44.906 AB + C --> AC + B "Fc1cccc(Cl)c1 + [OH-] ^{-1} --> Oc1cccc(c1)F + [Cl] ^{-1}"
485 -53.768 -50.819 -48.823 0.000 -48.823 AB + C --> AC + B "Fc1ccc(Cl)cc1 theory{pspw4} + [OH-] ^{-1} theory{pspw4} --> Oc1ccc(cc1)F theory{pspw4} + [Cl] ^{-1} theory{pspw4}"
476 -10.622 -7.848 -6.124 0.860 -5.264 AB + C --> AC + B "CCl + [OH] --> CO + [Cl]"
440 -56.107 -53.513 -51.755 15.151 -36.604 AB + C --> AC + B "CCCCl + [OH] ^{-1} --> CCCO + [Cl] ^{-1}"
All requests to Arrows were successful.
KEYWORDs -
reaction: :reaction
chinese_room: :chinese_room
molecule: :molecule
nmr: :nmr
predict: :predict
submitesmiles: :submitesmiles
nosubmitmissingesmiles
resubmitmissingesmiles
submitmachines: :submitmachines
useallentries
nomodelcorrect
eigenvalues: :eigenvalues
frequencies: :frequencies
nwoutput: :nwoutput
xyzfile: :xyzfile
alleigs: :alleigs
allfreqs: :allfreqs
reactionenumerate:
energytype:[erxn(gas) hrxn(gas) grxn(gas) delta_solvation grxn(aq)] :energytype
energytype:[kcal/mol kj/mol ev cm-1 ry hartree au] :energytype
tablereactions:
reaction: ... :reaction
reaction: ... :reaction
...
:tablereactions
tablemethods:
method: ... :method
method: ... :method
...
:tablemethods
:reactionenumerate
rotatebonds
xyzinput:
label: :label
xyzdata:
... xyz data ...
:xyzdata
:xyzinput
submitHf: :submitHf
nmrexp: :nmrexp
findreplace: old text | new text :findreplace
listnwjobs
fetchnwjob: :fetchnwjob
pushnwjob: :pushnwjob
printcsv: :printcsv
printeig: :printeig
printfreq: :printfreq
printxyz: :printxyz
printjobinfo: :printjobinfo
printnwout: :printnwout
badids: :badids
hup_string:
database:
table:
request_table:
listallesmiles
queuecheck
This software service and its documentation were developed at the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory, a multiprogram national laboratory, operated for the U.S. Department of Energy by Battelle under Contract Number DE-AC05-76RL01830. Support for this work was provided by the Department of Energy Office of Biological and Environmental Research, and Department of Defense environmental science and technology program (SERDP). THE SOFTWARE SERVICE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE SERVICE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE SERVICE.