 3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual
3D Periodic Editor 3D Molecular And Reaction Editor Expert Editor Microsoft Quantum Editor EMSL Aerosol Workshop Editor Manual Results from an EMSL Arrows Calculation
| EMSL Arrows is a revolutionary approach to materials and chemical simulations that uses NWChem and chemical computational databases to make materials and chemical modeling accessible via a broad spectrum of digital communications including posts to web APIs, social networks, and traditional email. |
Molecular modeling software has previously been extremely complex, making it prohibative to all but experts in the field, yet even experts can struggle to perform calculations. This service is designed to be used by experts and non-experts alike. Experts can carry out and keep track of large numbers of complex calculations with diverse levels of theories present in their workflows. Additionally, due to a streamlined and easy-to-use input, non-experts can carry out a wide variety of molecular modeling calculations previously not accessible to them.
The id(s) for emsiles = Cl[C]Cl theory{pspw4} xc{pbe0} basis{100.0 Ry} solvation_type{None} ^{0} mult{3} are: 61217
Use id=% instead of esmiles to print other entries.
mformula = C1Cl2
iupac = dichloromethane triplet radical
PubChem = 6344
PubChem LCSS = 6344
cas = 75-09-2
kegg = C02271 D02330
synonyms = DICHLOROMETHANE; Methylene chloride; 75-09-2; Methylene dichloride; Methane, dichloro-; Methylene bichloride; Methane dichloride; Solaesthin; Solmethine; Freon 30; Narkotil; Aerothene MM; Metylenu chlorek; Chlorure de methylene; Dichlormethan; Metaclen; Soleana VDA; CH2Cl2; Khladon 30; dichloro-methane; F 30 (chlorocarbon); RCRA waste number U080; NCI-C50102; R 30; dichlormethane; Methoklone; HCC 30; Salesthin; dichloro methane; Chlorodorm D; Methylene choride; UNII-588X2YUY0A; NSC 406122; UN 1593; F 30; Methylene chloride [NF]; MFCD00000881; Dichloromethane, HPLC Grade; METHYLENE CHLORIDE (DICHLOROMETHANE); 588X2YUY0A; CHEBI:15767; Methylene chloride (NF); Methylenum chloratum; R30 (refrigerant); Caswell No. 568; Metylenu chlorek [Polish]; HSDB 66; CCRIS 392; Chloride, Methylene; Chlorure de methylene [French]; Bichloride, Methylene; Dichloride, Methylene; EINECS 200-838-9; UN1593; DICHLOROMETHANE, NF; RCRA waste no. U080; DICHLOROMETHANE, ACS; EPA Pesticide Chemical Code 042004; BRN 1730800; dichioromethane; dichlormetane; dichloromeihane; dichlorometan; dichlorometane; dichloromethan; dichoromethane; dicloromethane; methylenchoride; metylenchloride; Dichloromethane, 99+%, extra pure, stabilized with amylene; Dichloromethane, 99+%, extra pure, stabilized with ethanol; Dichloromethane, 99.8%, for HPLC, stabilized with amylene; Methylenchlorid; Aerothene; Driverit; Nevolin; dichlor-methane; Dichloromethane, 99.5%, for analysis, stabilized with ethanol; Dichloromethane, 99.6%, ACS reagent, stabilized with amylene; Dichloromethane, 99.8%, for HPLC, stabilized with methanol; Dichloromethane, 99.8+%, for analysis, stabilized with amylene; Dichloromethane, 99.9%, Extra Dry, stabilized, AcroSeal(R); dichlorometliane; dichlorornethane; dicliloromethane; methylenchloride; methylenechlorid; methYIenechlorid; di-chloromethane; dichloromethane-; methlyenechloride; methylenechloride; AI3-01773; Dichloromethane, 99.8%, for spectroscopy, stabilized with amylene; Dichloromethane, for residue and pestic. anal., stab. with amylene; Dichloromethane, suitable for 5000 per JIS, for residue analysis; methlene chloride; methyene chloride; methylen chloride; methylene chlorie; methylene cloride; metylene chloride; Dichloromethane, 99.5%, for spectroscopy ACS, stabilized with amylene; Dichloromethane, 99.9%, for peptide synthesis, stabilized with amylene; mehtylene chloride; methlyene chloride; methylene,chloride; methylene-chloride; dichloro -methane; dichloro- methane; methylenedichloride; Distillex DS3; Dichloromethane, 99.8%, Extra Dry over Molecular Sieve, Stabilized, AcroSeal(R); Dichloromethane, ACS reagent, >=99.5%, contains 40-150 ppm amylene as stabilizer; M-clean D; methyl ene chloride; Dichloromethane, 99.9%, for residue analysis, for anal.of polyarom.hydrocarb., stab.with amylene; Dichloromethane (Methylene Chloride); MeCl2; DCM,SP Grade; methylene di chloride; N,N-methylenechloride; dichloromethane solution; Methylene chloride ACS; Cl2CH2; H2CCl2; DSSTox_CID_868; Dichloromethane, anhydrous; ACMC-1BFR1; Dichloromethane, for HPLC; EC 200-838-9; NCIMech_000221; WLN: G1G; DSSTox_RID_75836; DSSTox_GSID_20868; 4-01-00-00035 (Beilstein Handbook Reference); Dichloromethane, >=99.9%; CHEMBL45967; Methylene Chloride (Recovered); Dichloromethane, AR, >=99%; Dichloromethane (Peptide Grade); DTXSID0020868; DTXSID60166893; Dichloromethane Reagent Grade ACS; METHYLENE CHLORIDE (13C); Dichloromethane, LR, >=99.5%; Dichloromethane, purification grade; Dichloromethane, analytical standard; Dichloromethane, Environmental Grade; Tox21_202526; ANW-42404; NSC406122; STL264204; AKOS009031498; Dichloromethane [UN1593] [Poison]; Dichloromethane, ACS reagent, 99.5%; MCULE-1114735075; NSC-406122; CAS-75-09-2; Dichloromethane, for HPLC, >=99.7%; Dichloromethane GC, for residue analysis; Dichloromethane, Spectrophotometric Grade; NCGC00091504-01; NCGC00260075-01; Dichloromethane 100 microg/mL in Methanol; Dichloromethane, suitable for PCB analysis; Dichloromethane 1000 microg/mL in Methanol; D0529; D3478; FT-0624716; FT-0624717; M0629; Dichloromethane, for HPLC, >=99.8% (GC); Dichloromethane, SAJ first grade, >=99.0%; Dichloromethane, Selectophore(TM), >=99.5%; C02271; D02330; Dichloromethane, analytical standard, stabilized; Dichloromethane, JIS special grade, >=99.0%; L023970; Q421748; Q425210; J-610006; Dichloromethane, 99%, stab. with ca. 50ppm amylene; Methylene Chloride HPLC grade Stabilized with Amylene; Dichloromethane solution, contains 10 % (v/v) methanol; Dichloromethane, glass distilled HRGC/HPLC trace grade; Dichloromethane, TLC high-purity grade, >=99.8% (GC); Dichloromethane HPLC, UV/IR, min. 99.9%, isocratic grade; Dichloromethane, special, 99.9%, contains 40-60 ppm Amylene; Dichloromethane, for HPLC, >=99.8%, contains amylene as stabilizer; Dichloromethane, Selectophore(TM), >=99.5% (GC), inhibitor-free; Dichloromethane, suitable for 300 per JIS, for residue analysis; Dichloromethane, technical grade, 95%, contains 40-60 ppm Amylene; Methylene chloride, European Pharmacopoeia (EP) Reference Standard; Dichloromethane, puriss. p.a., ACS reagent, reag. ISO, >=99.9% (GC); Dichloromethane, UV HPLC spectroscopic, 99.9%, contains 40-60 ppm Amylene; Dichloromethane solution, 10 % (v/v) in methanol, 1 % (v/v) in ammonium hydroxide; Dichloromethane solution, certified reference material, 200 mug/mL in methanol; Dichloromethane solution, certified reference material, 5000 mug/mL in methanol; Dichloromethane, 99.9%, Extra Dry, stabilized, AcroSeal(R), package of 4x25ML bottles; Dichloromethane, ACS reagent, >=99.5%, contains 50 ppm amylene as stabilizer; Dichloromethane, anhydrous, >=99.8%, contains 40-150 ppm amylene as stabilizer; Dichloromethane, anhydrous, contains 40-150 ppm amylene as stabilizer, ZerO2(TM), >=99.8%; Dichloromethane, biotech. grade, 99.9%, contains 40-150 ppm amylene as stabilizer; Dichloromethane, contains 40-150 ppm amylene as stabilizer, ACS reagent, >=99.5%; Dichloromethane, for HPLC, >=99.9%, contains 40-150 ppm amylene as stabilizer; Dichloromethane, puriss., meets analytical specification of Ph.??Eur., NF, >=99% (GC); Dichloromethane, suitable for 1000 per JIS, >=99.5%, for residue analysis; Methylene Chloride, Pharmaceutical Secondary Standard; Certified Reference Material; Dichloromethane, >=99.9%, capillary GC grade, suitable for environmental analysis, contains amylene as stabilizer; Dichloromethane, ACS spectrophotometric grade, >=99.5%, contains 50-150 ppm amylene as stabilizer; Dichloromethane, HPLC Plus, for HPLC, GC, and residue analysis, >=99.9%, contains 50-150 ppm amylene as stabilizer; Dichloromethane, Laboratory Reagent, >=99.9% (without stabilizer, GC), contains 0.1-0.4% ethanol as stabilizer; Dichloromethane, p.a., ACS reagent, reag. ISO, reag. Ph. Eur., 99.8%, contains 40-60 ppm Amylene; Dichloromethane, puriss. p.a., ACS reagent, reag. ISO, dried, >=99.8% (GC), <=0.001% water; M.C; Residual Solvent Class 2 - Methylene chloride, United States Pharmacopeia (USP) Reference Standard
Search Links to Other Online Resources (may not be available):
- Google Structure Search
- EPA CompTox Database
- Chemical Entities of Biological Interest (ChEBI)
- NIH ChemIDplus - A TOXNET DATABASE
- The Human Metabolome Database (HMDB)
- OECD eChemPortal
- Google Scholar
+==================================================+
|| Molecular Calculation ||
+==================================================+
Id = 61217
NWOutput = Link to NWChem Output (download)
Datafiles:
lumo-beta.cube-553684-2021-7-17-17:37:2 (download)
lumo-alpha.cube-553684-2021-7-17-17:37:2 (download)
density.cube-553684-2021-7-17-17:37:2 (download)
homo-alpha.cube-553684-2021-7-17-17:37:2 (download)
homo-beta.cube-553684-2021-7-17-17:37:2 (download)
image_resset: api/image_reset/61217
Calculation performed by Eric Bylaska - arrow15.emsl.pnl.gov
Numbers of cpus used for calculation = 32
Calculation walltime = 26575.900000 seconds (0 days 7 hours 22 minutes 55 seconds)
+----------------+
| Energetic Data |
+----------------+
Id = 61217
iupac = dichloromethane triplet radical
mformula = C1Cl2
inchi = InChI=1S/CCl2/c2-1-3
inchikey = PFBUKDPBVNJDEW-UHFFFAOYSA-N
esmiles = Cl[C]Cl theory{pspw4} xc{pbe0} basis{100.0 Ry} solvation_type{None} ^{0} mult{3}
calculation_type = ov
theory = pspw4
xc = pbe0
basis = 100.0 Ry
charge,mult = 0 3
energy = -35.788517 Hartrees
enthalpy correct.= 0.009052 Hartrees
entropy = 63.946 cal/mol-K
solvation energy = 0.000 kcal/mol solvation_type = None
Sitkoff cavity dispersion = 1.801 kcal/mol
Honig cavity dispersion = 4.704 kcal/mol
ASA solvent accesible surface area = 188.149 Angstrom2
ASA solvent accesible volume = 196.996 Angstrom3
+-----------------+
| Structural Data |
+-----------------+
JSmol: an open-source HTML5 viewer for chemical structures in 3D
Number of Atoms = 3
Units are Angstrom for bonds and degrees for angles
Type I J K L M Value
----------- ----- ----- ----- ----- ----- ----------
1 Stretch C1 Cl2 1.65244
2 Stretch C1 Cl3 1.65241
3 Bend Cl2 C1 Cl3 128.96832
+-----------------+
| Eigenvalue Data |
+-----------------+
Id = 61217
iupac = dichloromethane triplet radical
mformula = C1Cl2
InChI = InChI=1S/CCl2/c2-1-3
smiles = Cl[C]Cl
esmiles = Cl[C]Cl theory{pspw4} xc{pbe0} basis{100.0 Ry} solvation_type{None} ^{0} mult{3}
theory = pspw4
xc = pbe0
basis = 100.0 Ry
charge = 0
mult = 3
solvation_type = None
twirl webpage = TwirlMol Link
image webpage = GIF Image Link
Unrestricted Eigevalue Spectra
alpha beta
- - - - -- 0.94 eV -- -- -- - 0.89 eV
---------- ----------
---- ----LUMO= -0.37 eV ----------
----------
----------LUMO= -2.86 eV
HOMO= -6.22 eV++++++++++
++++++++++
++++++++++ HOMO= -9.67 eV++++ ++++
++++++++++
++++ ++++
++++ ++++
++++++++++
++++++++++
++++++++++
++++++++++
++++++++++
++++++++++
-25.86 eV++++++++++
-26.53 eV++++++++++

spin eig occ ---------------------------- alpha -6.22 1.00 alpha -7.99 1.00 alpha -9.92 1.00 alpha -10.28 1.00 alpha -12.43 1.00 alpha -12.44 1.00 alpha -13.93 1.00 alpha -17.57 1.00 alpha -24.64 1.00 alpha -26.53 1.00 alpha 0.94 0.00 alpha 0.91 0.00 alpha 0.78 0.00 alpha 0.78 0.00 alpha 0.74 0.00 alpha 0.51 0.00 alpha -0.19 0.00 alpha -0.37 0.00 beta -9.67 1.00 beta -9.79 1.00 beta -11.18 1.00 beta -11.27 1.00 beta -13.36 1.00 beta -16.10 1.00 beta -24.31 1.00 beta -25.86 1.00 beta 0.89 0.00 beta 0.86 0.00 beta 0.85 0.00 beta 0.66 0.00 beta 0.28 0.00 beta -0.19 0.00 beta -1.29 0.00 beta -2.86 0.00
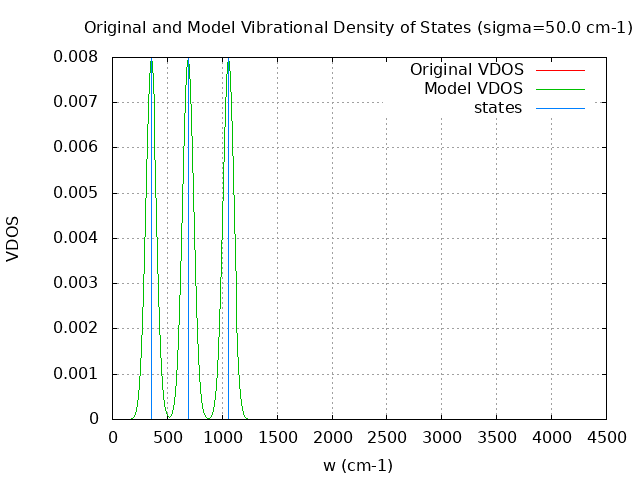
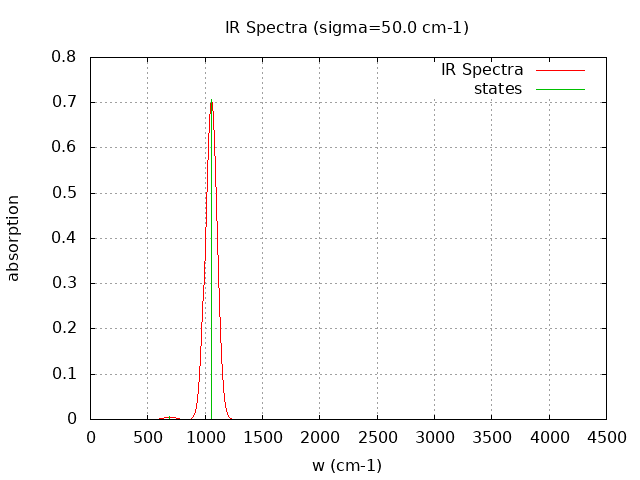
+----------------------------------------+ | Vibrational Density of States Analysis | +----------------------------------------+ Total number of frequencies = 9 Total number of negative frequencies = 0 Number of lowest frequencies = 1 (frequency threshold = 500 ) Exact dos norm = 3.000 Generating vibrational DOS Generating model vibrational DOS to have a proper norm 10.00 3.00 1.00 3.00 50.00 3.00 1.00 3.00 100.00 3.00 1.00 3.00 Generating IR Spectra Writing vibrational density of states (DOS) to vdos.dat Writing model vibrational density of states (DOS_FIXED) to vdos-model.dat Writing IR spectra to irdos.dat Temperature= 298.15 zero-point correction to energy = 2.993 kcal/mol ( 0.004770) vibrational contribution to enthalpy correction = 3.311 kcal/mol ( 0.005277) vibrational contribution to Entropy = 1.554 cal/mol-k hindered rotor enthalpy correction = 0.000 kcal/mol ( 0.000000) hindered rotor entropy correction = 0.000 cal/mol-k
DOS sigma = 10.000000
- vibrational DOS enthalpy correction = 0.005277 kcal/mol ( 3.311 kcal/mol)
- model vibrational DOS enthalpy correction = 0.005273 kcal/mol ( 3.309 kcal/mol)
- vibrational DOS Entropy = 0.000002 ( 1.556 cal/mol-k)
- model vibrational DOS Entropy = 0.000002 ( 1.551 cal/mol-k)
- original gas Energy = -35.788517 (-22457.633 kcal/mol)
- original gas Enthalpy = -35.779465 (-22451.953 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -35.779465 (-22451.953 kcal/mol, delta= 0.000)
- model DOS gas Enthalpy = -35.779469 (-22451.956 kcal/mol, delta= -0.002)
- original gas Entropy = 0.000102 ( 63.946 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000102 ( 63.947 cal/mol-k,delta= 0.001)
- model DOS gas Entropy = 0.000102 ( 63.943 cal/mol-k,delta= -0.003)
- original gas Free Energy = -35.809848 (-22471.019 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -35.809848 (-22471.019 kcal/mol, delta= -0.000)
- model DOS gas Free Energy = -35.809851 (-22471.020 kcal/mol, delta= -0.002)
- original sol Free Energy = -35.809848 (-22471.019 kcal/mol)
- unadjusted DOS sol Free Energy = -35.809848 (-22471.019 kcal/mol)
- model DOS sol Free Energy = -35.809851 (-22471.020 kcal/mol)
DOS sigma = 50.000000
- vibrational DOS enthalpy correction = 0.005282 kcal/mol ( 3.315 kcal/mol)
- model vibrational DOS enthalpy correction = 0.005282 kcal/mol ( 3.315 kcal/mol)
- vibrational DOS Entropy = 0.000003 ( 1.586 cal/mol-k)
- model vibrational DOS Entropy = 0.000003 ( 1.586 cal/mol-k)
- original gas Energy = -35.788517 (-22457.633 kcal/mol)
- original gas Enthalpy = -35.779465 (-22451.953 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -35.779460 (-22451.950 kcal/mol, delta= 0.004)
- model DOS gas Enthalpy = -35.779460 (-22451.950 kcal/mol, delta= 0.004)
- original gas Entropy = 0.000102 ( 63.946 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000102 ( 63.977 cal/mol-k,delta= 0.031)
- model DOS gas Entropy = 0.000102 ( 63.977 cal/mol-k,delta= 0.031)
- original gas Free Energy = -35.809848 (-22471.019 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -35.809857 (-22471.024 kcal/mol, delta= -0.006)
- model DOS gas Free Energy = -35.809857 (-22471.024 kcal/mol, delta= -0.006)
- original sol Free Energy = -35.809848 (-22471.019 kcal/mol)
- unadjusted DOS sol Free Energy = -35.809857 (-22471.024 kcal/mol)
- model DOS sol Free Energy = -35.809857 (-22471.024 kcal/mol)
DOS sigma = 100.000000
- vibrational DOS enthalpy correction = 0.005299 kcal/mol ( 3.325 kcal/mol)
- model vibrational DOS enthalpy correction = 0.005299 kcal/mol ( 3.325 kcal/mol)
- vibrational DOS Entropy = 0.000003 ( 1.691 cal/mol-k)
- model vibrational DOS Entropy = 0.000003 ( 1.692 cal/mol-k)
- original gas Energy = -35.788517 (-22457.633 kcal/mol)
- original gas Enthalpy = -35.779465 (-22451.953 kcal/mol, delta= 0.000)
- unajusted DOS gas Enthalpy = -35.779443 (-22451.939 kcal/mol, delta= 0.014)
- model DOS gas Enthalpy = -35.779442 (-22451.939 kcal/mol, delta= 0.014)
- original gas Entropy = 0.000102 ( 63.946 cal/mol-k,delta= 0.000)
- unajusted DOS gas Entropy = 0.000102 ( 64.083 cal/mol-k,delta= 0.137)
- model DOS gas Entropy = 0.000102 ( 64.083 cal/mol-k,delta= 0.137)
- original gas Free Energy = -35.809848 (-22471.019 kcal/mol, delta= 0.000)
- unadjusted DOS gas Free Energy = -35.809891 (-22471.045 kcal/mol, delta= -0.027)
- model DOS gas Free Energy = -35.809890 (-22471.045 kcal/mol, delta= -0.027)
- original sol Free Energy = -35.809848 (-22471.019 kcal/mol)
- unadjusted DOS sol Free Energy = -35.809891 (-22471.045 kcal/mol)
- model DOS sol Free Energy = -35.809890 (-22471.045 kcal/mol)
Normal Mode Frequency (cm-1) IR Intensity (arbitrary)
----------- ---------------- ------------------------
1 -0.000 0.015
2 -0.000 0.162
3 -0.000 0.015
4 0.000 0.527
5 0.000 0.030
6 0.000 0.027
7 351.060 0.034
8 689.850 0.693
9 1053.840 88.497
No Hindered Rotor Data
+-------------------------------------+
| Reactions Contained in the Database |
+-------------------------------------+
Reactions Containing INCHIKEY = PFBUKDPBVNJDEW-UHFFFAOYSA-N
Reactionid Erxn(gas) Hrxn(gas) Grxn(gas) delta_Solv Grxn(aq) ReactionType Reaction
19949 -86.190 -83.218 -81.155 7.826 -73.329 AB + C --> AC + B "[C](Cl)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> [C](Cl)O xc{m06-2x} + [Cl-] xc{m06-2x}"
19794 -66.362 -63.875 -62.017 8.437 -53.581 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{m06-2x} + [OH-] xc{m06-2x} --> [C](Cl)O mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
19310 -63.910 -60.274 -48.416 3.174 -45.241 AB + C --> ACB "[C](Cl)Cl + O --> C(Cl)(Cl)O"
19309 -63.910 -60.274 -48.416 3.174 -45.241 AB + C --> ACB "[C](Cl)Cl + O --> C(Cl)(Cl)O"
17252 365.683 359.165 351.033 -315.048 35.985 AB --> A + B "Cl[CH]Cl mult{2} --> [Cl]=C=[Cl] ^{-1} mult{2} + [H] ^{1}"
17251 365.683 359.165 351.033 -315.048 35.985 AB --> A + B "Cl[CH]Cl mult{2} --> [Cl]=C=[Cl] ^{-1} mult{2} + [H] ^{1}"
17208 -21.902 -22.965 -31.438 -81.432 -14.271 AB --> A + B "Cl[C](Cl)Cl mult{2} + [SHE] --> Cl[C]Cl + [Cl] ^{-1}"
17207 -21.902 -22.965 -31.438 -81.432 -14.271 AB --> A + B "Cl[C](Cl)Cl mult{2} + [SHE] --> Cl[C]Cl + [Cl] ^{-1}"
16964 -61.600 -57.971 -46.116 3.154 -42.962 AB + C --> ACB "[C](Cl)Cl + O --> C(Cl)(Cl)O"
16963 -61.600 -57.971 -46.116 3.154 -42.962 AB + C --> ACB "[C](Cl)Cl + O --> C(Cl)(Cl)O"
15631 423.086 417.357 409.980 -256.956 54.424 AB --> A + B "Cl[CH]Cl mult{2} theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [H+] theory{ccsd(t)} + SHE theory{ccsd(t)}"
15630 423.086 417.357 409.980 -256.956 54.424 AB --> A + B "Cl[CH]Cl mult{2} theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [H+] theory{ccsd(t)} + SHE theory{ccsd(t)}"
15625 -30.955 -29.704 -27.839 -8.539 -36.378 AB + C --> AC + B "[C](Cl)Cl mult{3} theory{ccsd(t)} + [SH-] theory{ccsd(t)} --> [C](Cl)S mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
15569 -59.758 -57.453 -55.582 13.777 -41.806 AB + C --> AC + B "[C](Cl)Cl mult{3} theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> [C](Cl)O mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
15568 50.007 50.075 42.516 -26.204 16.312 AB --> A + B "[C-](Cl)(Cl)Cl theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
15567 50.007 50.075 42.516 -26.204 16.312 AB --> A + B "[C-](Cl)(Cl)Cl theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
15489 -2.469 -2.619 -9.204 -29.094 -38.298 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
15488 -2.469 -2.619 -9.204 -29.094 -38.298 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
15487 2.025 1.940 -4.536 -30.499 -35.035 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
15486 2.025 1.940 -4.536 -30.499 -35.035 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
15485 15.886 15.970 9.461 -27.730 -18.269 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
15484 15.886 15.970 9.461 -27.730 -18.269 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
15483 16.510 16.652 10.264 -29.056 -18.791 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
15482 16.510 16.652 10.264 -29.056 -18.791 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
15463 0.620 0.451 -7.486 -34.569 -42.055 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
15462 0.620 0.451 -7.486 -34.569 -42.055 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
15461 5.949 5.975 -1.134 0.000 -1.134 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} theory{pspw4} --> [C](Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
15460 5.949 5.975 -1.134 0.000 -1.134 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} theory{pspw4} --> [C](Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
15459 20.188 20.272 12.457 -33.388 -20.931 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
15458 20.188 20.272 12.457 -33.388 -20.931 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
15457 20.222 20.573 13.615 0.000 13.615 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
15456 20.222 20.573 13.615 0.000 13.615 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
15440 -28.489 -27.219 -25.401 -8.618 -34.020 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{m06-2x} + [SH-] xc{m06-2x} --> [C](Cl)S mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
15439 -28.496 -27.229 -25.371 -8.518 -33.889 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{pbe0} + [SH-] xc{pbe0} --> [C](Cl)S mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
15438 -27.647 -26.562 -24.701 -8.348 -33.049 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{pbe} + [SH-] xc{pbe} --> [C](Cl)S mult{3} xc{pbe} + [Cl-] xc{pbe}"
15437 -29.143 -27.891 -26.027 -8.539 -34.566 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{b3lyp} + [SH-] xc{b3lyp} --> [C](Cl)S mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
15436 -28.220 -26.896 -24.880 0.000 -24.880 AB + C --> AC + B "[C](Cl)Cl mult{3} theory{pspw4} + [SH-] theory{pspw4} --> [C](Cl)S mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
15435 1.305 1.241 -5.139 -25.935 -31.074 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{b3lyp} --> [C](Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
15434 1.305 1.241 -5.139 -25.935 -31.074 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{b3lyp} --> [C](Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
15417 19.514 19.663 13.378 -24.623 -11.244 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
15416 19.514 19.663 13.378 -24.623 -11.244 AB --> A + B "[C-](Cl)(Cl)Cl mult{3} xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
15371 51.358 51.355 43.467 -25.835 17.632 AB --> A + B "[C-](Cl)(Cl)Cl xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
15370 51.358 51.355 43.467 -25.835 17.632 AB --> A + B "[C-](Cl)(Cl)Cl xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
15369 46.807 46.833 39.035 -25.757 13.278 AB --> A + B "[C-](Cl)(Cl)Cl xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
15368 46.807 46.833 39.035 -25.757 13.278 AB --> A + B "[C-](Cl)(Cl)Cl xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
15367 52.738 52.836 45.336 -26.166 19.170 AB --> A + B "[C-](Cl)(Cl)Cl xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
15366 52.738 52.836 45.336 -26.166 19.170 AB --> A + B "[C-](Cl)(Cl)Cl xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
15365 46.144 46.212 38.653 -26.204 12.449 AB --> A + B "[C-](Cl)(Cl)Cl xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
15364 46.144 46.212 38.653 -26.204 12.449 AB --> A + B "[C-](Cl)(Cl)Cl xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
15363 46.103 46.197 38.452 0.000 38.452 AB --> A + B "[C-](Cl)(Cl)Cl theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
15362 46.103 46.197 38.452 0.000 38.452 AB --> A + B "[C-](Cl)(Cl)Cl theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
15296 423.308 417.458 410.062 -256.755 54.707 AB --> A + B "Cl[CH]Cl xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + SHE xc{m06-2x} + [H+] xc{m06-2x}"
15295 423.308 417.458 410.062 -256.755 54.707 AB --> A + B "Cl[CH]Cl xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + SHE xc{m06-2x} + [H+] xc{m06-2x}"
15294 423.304 417.544 410.169 -256.856 54.712 AB --> A + B "Cl[CH]Cl xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + SHE xc{pbe0} + [H+] xc{pbe0}"
15293 423.304 417.544 410.169 -256.856 54.712 AB --> A + B "Cl[CH]Cl xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + SHE xc{pbe0} + [H+] xc{pbe0}"
15292 421.869 416.366 409.027 -257.097 53.330 AB --> A + B "Cl[CH]Cl xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + SHE xc{pbe} + [H+] xc{pbe}"
15291 421.869 416.366 409.027 -257.097 53.330 AB --> A + B "Cl[CH]Cl xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + SHE xc{pbe} + [H+] xc{pbe}"
15290 425.922 420.192 412.815 -256.956 57.260 AB --> A + B "Cl[CH]Cl xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + SHE xc{b3lyp} + [H+] xc{b3lyp}"
15289 425.922 420.192 412.815 -256.956 57.260 AB --> A + B "Cl[CH]Cl xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + SHE xc{b3lyp} + [H+] xc{b3lyp}"
15288 421.059 415.473 408.130 0.000 309.530 AB --> A + B "Cl[CH]Cl theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + SHE theory{pspw4} + [H+] theory{pspw4}"
15287 421.059 415.473 408.130 0.000 309.530 AB --> A + B "Cl[CH]Cl theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + SHE theory{pspw4} + [H+] theory{pspw4}"
15257 -66.362 -63.875 -62.018 14.287 -47.731 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{m06-2x} + [OH-] xc{m06-2x} --> [C](Cl)O mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
15256 -63.226 -60.842 -58.993 13.657 -45.336 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{pbe0} + [OH-] xc{pbe0} --> [C](Cl)O mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
15255 -58.740 -56.627 -54.754 13.398 -41.356 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{pbe} + [OH-] xc{pbe} --> [C](Cl)O mult{3} xc{pbe} + [Cl-] xc{pbe}"
15254 -62.099 -59.794 -57.923 13.777 -44.147 AB + C --> AC + B "[C](Cl)Cl mult{3} xc{b3lyp} + [OH-] xc{b3lyp} --> [C](Cl)O mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
15142 -64.496 -60.622 -48.681 0.000 -48.681 AB + C --> ACB "[C](Cl)Cl theory{pspw4} + O theory{pspw4} --> C(Cl)(Cl)O theory{pspw4}"
15141 -64.496 -60.622 -48.681 0.000 -48.681 AB + C --> ACB "[C](Cl)Cl theory{pspw4} + O theory{pspw4} --> C(Cl)(Cl)O theory{pspw4}"
15041 -70.498 -66.730 -54.749 3.173 -51.576 AB + C --> ACB "[C](Cl)Cl xc{m06-2x} + O xc{m06-2x} --> C(Cl)(Cl)O xc{m06-2x}"
15040 -70.498 -66.730 -54.749 3.173 -51.576 AB + C --> ACB "[C](Cl)Cl xc{m06-2x} + O xc{m06-2x} --> C(Cl)(Cl)O xc{m06-2x}"
15039 -70.038 -66.423 -54.528 3.234 -51.294 AB + C --> ACB "[C](Cl)Cl xc{pbe0} + O xc{pbe0} --> C(Cl)(Cl)O xc{pbe0}"
15038 -70.038 -66.423 -54.528 3.234 -51.294 AB + C --> ACB "[C](Cl)Cl xc{pbe0} + O xc{pbe0} --> C(Cl)(Cl)O xc{pbe0}"
15037 -65.164 -61.772 -49.951 3.023 -46.928 AB + C --> ACB "[C](Cl)Cl xc{pbe} + O xc{pbe} --> C(Cl)(Cl)O xc{pbe}"
15036 -65.164 -61.772 -49.951 3.023 -46.928 AB + C --> ACB "[C](Cl)Cl xc{pbe} + O xc{pbe} --> C(Cl)(Cl)O xc{pbe}"
15026 92.331 86.247 78.775 0.875 79.651 AB --> A + B "[CH](Cl)Cl xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [H] xc{m06-2x}"
15025 92.331 86.247 78.775 0.875 79.651 AB --> A + B "[CH](Cl)Cl xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [H] xc{m06-2x}"
15024 94.344 88.357 80.894 0.695 81.588 AB --> A + B "[CH](Cl)Cl xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [H] xc{pbe0}"
15023 94.344 88.357 80.894 0.695 81.588 AB --> A + B "[CH](Cl)Cl xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [H] xc{pbe0}"
15022 88.663 82.908 75.446 0.717 76.163 AB --> A + B "[CH](Cl)Cl xc{pbe} --> [C](Cl)Cl xc{pbe} + [H] xc{pbe}"
15021 88.663 82.908 75.446 0.717 76.163 AB --> A + B "[CH](Cl)Cl xc{pbe} --> [C](Cl)Cl xc{pbe} + [H] xc{pbe}"
15015 -63.910 -60.276 -48.415 3.075 -45.340 AB + C --> ACB "[C](Cl)Cl + O --> C(Cl)(Cl)O"
15014 -63.910 -60.276 -48.415 3.075 -45.340 AB + C --> ACB "[C](Cl)Cl + O --> C(Cl)(Cl)O"
15010 92.542 86.599 79.127 0.726 79.854 AB --> A + B "[CH](Cl)Cl --> [C](Cl)Cl + [H]"
15009 92.542 86.599 79.127 0.726 79.854 AB --> A + B "[CH](Cl)Cl --> [C](Cl)Cl + [H]"
14738 -78.693 -75.879 -73.816 13.035 -60.781 AB + C --> AC + B "[C](Cl)Cl theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> [C](Cl)O theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
11670 -82.074 -79.238 -77.176 13.067 -64.109 AB + C --> AC + B "[C](Cl)Cl xc{pbe0} + [OH-] xc{pbe0} --> [C](Cl)O xc{pbe0} + [Cl-] xc{pbe0}"
8010 199.671 199.526 191.872 -236.037 54.435 AB --> A + B "Cl[C](Cl)Cl ^{-1} + [SHE] --> Cl[C]Cl + [Cl] ^{-2} mult{2}"
8009 199.671 199.526 191.872 -236.037 54.435 AB --> A + B "Cl[C](Cl)Cl ^{-1} + [SHE] --> Cl[C]Cl + [Cl] ^{-2} mult{2}"
7969 371.510 369.871 360.737 -157.131 203.606 AB --> A + B "Cl[C](Cl)Cl mult{2} --> Cl[C]Cl ^{-1} mult{2} + [Cl] ^{1}"
7968 371.510 369.871 360.737 -157.131 203.606 AB --> A + B "Cl[C](Cl)Cl mult{2} --> Cl[C]Cl ^{-1} mult{2} + [Cl] ^{1}"
7238 31.347 31.099 23.018 0.000 23.018 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} --> [C](Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7237 31.347 31.099 23.018 0.000 23.018 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} xc{pbe0} --> [C](Cl)Cl theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7232 3.554 2.605 -5.868 0.000 92.732 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)Cl mult{3} theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7231 3.554 2.605 -5.868 0.000 92.732 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} xc{pbe0} + SHE theory{pspw4} xc{pbe0} --> [C](Cl)Cl mult{3} theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7193 -78.693 -75.879 -73.816 13.115 -60.700 AB + C --> AC + B "[C](Cl)Cl theory{ccsd(t)} + [OH-] theory{ccsd(t)} --> [C](Cl)O theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7190 30.204 30.059 22.404 -27.517 -5.113 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} --> [C](Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7189 30.204 30.059 22.404 -27.517 -5.113 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{ccsd(t)} --> [C](Cl)Cl theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7180 4.597 3.744 -4.666 0.000 93.934 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} + SHE theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
7179 4.597 3.744 -4.666 0.000 93.934 AB --> A + B "[C](Cl)(Cl)Cl theory{pspw4} + SHE theory{pspw4} --> [C](Cl)Cl mult{3} theory{pspw4} + [Cl-] theory{pspw4}"
7158 -78.073 -74.990 -72.958 0.000 -72.958 AB + C --> AC + B "[C](Cl)Cl theory{pspw4} xc{pbe0} + [OH-] theory{pspw4} xc{pbe0} --> [C](Cl)O theory{pspw4} xc{pbe0} + [Cl-] theory{pspw4} xc{pbe0}"
7157 -69.940 -67.084 -65.023 0.000 -65.023 AB + C --> AC + B "[C](Cl)Cl theory{pspw4} + [OH-] theory{pspw4} --> [C](Cl)O theory{pspw4} + [Cl-] theory{pspw4}"
7156 -86.191 -83.218 -81.155 13.676 -67.479 AB + C --> AC + B "[C](Cl)Cl xc{m06-2x} + [OH-] xc{m06-2x} --> [C](Cl)O xc{m06-2x} + [Cl-] xc{m06-2x}"
7155 -82.074 -79.238 -77.176 13.127 -64.049 AB + C --> AC + B "[C](Cl)Cl xc{pbe0} + [OH-] xc{pbe0} --> [C](Cl)O xc{pbe0} + [Cl-] xc{pbe0}"
7154 -76.061 -73.503 -71.422 12.644 -58.777 AB + C --> AC + B "[C](Cl)Cl xc{pbe} + [OH-] xc{pbe} --> [C](Cl)O xc{pbe} + [Cl-] xc{pbe}"
7153 -80.758 -77.944 -75.881 13.035 -62.846 AB + C --> AC + B "[C](Cl)Cl xc{b3lyp} + [OH-] xc{b3lyp} --> [C](Cl)O xc{b3lyp} + [Cl-] xc{b3lyp}"
7141 33.003 32.766 24.802 -27.199 -2.397 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
7140 33.003 32.766 24.802 -27.199 -2.397 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{m06-2x} --> [C](Cl)Cl xc{m06-2x} + [Cl-] xc{m06-2x}"
7139 32.322 32.121 24.235 -27.200 -2.965 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
7138 32.322 32.121 24.235 -27.200 -2.965 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe0} --> [C](Cl)Cl xc{pbe0} + [Cl-] xc{pbe0}"
7137 33.170 33.015 25.392 -27.347 -1.954 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
7136 33.170 33.015 25.392 -27.347 -1.954 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{pbe} --> [C](Cl)Cl xc{pbe} + [Cl-] xc{pbe}"
7119 5.281 4.430 -3.948 -80.119 14.533 AB --> A + B "[C](Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7118 5.281 4.430 -3.948 -80.119 14.533 AB --> A + B "[C](Cl)(Cl)Cl theory{ccsd(t)} + SHE theory{ccsd(t)} --> [C](Cl)Cl mult{3} theory{ccsd(t)} + [Cl-] theory{ccsd(t)}"
7117 1.670 0.749 -7.693 -80.697 10.210 AB --> A + B "[C](Cl)(Cl)Cl xc{m06-2x} + SHE xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
7116 1.670 0.749 -7.693 -80.697 10.210 AB --> A + B "[C](Cl)(Cl)Cl xc{m06-2x} + SHE xc{m06-2x} --> [C](Cl)Cl mult{3} xc{m06-2x} + [Cl-] xc{m06-2x}"
7099 31.830 31.599 23.703 0.000 23.703 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} --> [C](Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
7098 31.830 31.599 23.703 0.000 23.703 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} theory{pspw4} --> [C](Cl)Cl theory{pspw4} + [Cl-] theory{pspw4}"
7097 27.935 27.790 20.136 -27.517 -7.381 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} --> [C](Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7096 27.935 27.790 20.136 -27.517 -7.381 AB --> A + B "[C](Cl)(Cl)Cl ^{-1} xc{b3lyp} --> [C](Cl)Cl xc{b3lyp} + [Cl-] xc{b3lyp}"
7070 3.231 2.327 -6.092 -80.708 11.800 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe0} + SHE xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
7069 3.231 2.327 -6.092 -80.708 11.800 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe0} + SHE xc{pbe0} --> [C](Cl)Cl mult{3} xc{pbe0} + [Cl-] xc{pbe0}"
7068 4.846 4.051 -4.264 -79.950 14.386 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe} + SHE xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
7067 4.846 4.051 -4.264 -79.950 14.386 AB --> A + B "[C](Cl)(Cl)Cl xc{pbe} + SHE xc{pbe} --> [C](Cl)Cl mult{3} xc{pbe} + [Cl-] xc{pbe}"
7066 -3.693 -4.543 -12.921 -80.119 5.559 AB --> A + B "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
7065 -3.693 -4.543 -12.921 -80.119 5.559 AB --> A + B "[C](Cl)(Cl)Cl xc{b3lyp} + SHE xc{b3lyp} --> [C](Cl)Cl mult{3} xc{b3lyp} + [Cl-] xc{b3lyp}"
749 -3.693 -4.543 -12.921 -80.119 -93.041 AB --> A + B "Cl[C](Cl)Cl xyzdata{C -4.00022 0.58207 0.96997 | Cl -2.30229 0.97442 1.34210 | Cl -4.34205 -1.12742 1.31115 | Cl -4.38438 0.98070 -0.71795} --> [Cl] xyzdata{Cl 2.75521 1.83484 -0.24354} ^{-1} + Cl[C]Cl xyzdata{C 3.04352 -2.52273 -0.25387 | Cl 1.77592 -1.56644 -1.04794 | Cl 3.94514 -1.50287 0.88596} mult{3}"
All requests to Arrows were successful.
KEYWORDs -
reaction: :reaction
chinese_room: :chinese_room
molecule: :molecule
nmr: :nmr
predict: :predict
submitesmiles: :submitesmiles
nosubmitmissingesmiles
resubmitmissingesmiles
submitmachines: :submitmachines
useallentries
nomodelcorrect
eigenvalues: :eigenvalues
frequencies: :frequencies
nwoutput: :nwoutput
xyzfile: :xyzfile
alleigs: :alleigs
allfreqs: :allfreqs
reactionenumerate:
energytype:[erxn(gas) hrxn(gas) grxn(gas) delta_solvation grxn(aq)] :energytype
energytype:[kcal/mol kj/mol ev cm-1 ry hartree au] :energytype
tablereactions:
reaction: ... :reaction
reaction: ... :reaction
...
:tablereactions
tablemethods:
method: ... :method
method: ... :method
...
:tablemethods
:reactionenumerate
rotatebonds
xyzinput:
label: :label
xyzdata:
... xyz data ...
:xyzdata
:xyzinput
submitHf: :submitHf
nmrexp: :nmrexp
findreplace: old text | new text :findreplace
listnwjobs
fetchnwjob: :fetchnwjob
pushnwjob: :pushnwjob
printcsv: :printcsv
printeig: :printeig
printfreq: :printfreq
printxyz: :printxyz
printjobinfo: :printjobinfo
printnwout: :printnwout
badids: :badids
hup_string:
database:
table:
request_table:
listallesmiles
queuecheck
This software service and its documentation were developed at the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory, a multiprogram national laboratory, operated for the U.S. Department of Energy by Battelle under Contract Number DE-AC05-76RL01830. Support for this work was provided by the Department of Energy Office of Biological and Environmental Research, and Department of Defense environmental science and technology program (SERDP). THE SOFTWARE SERVICE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE SERVICE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE SERVICE.